How to parse efficiently with mapping annotations¶
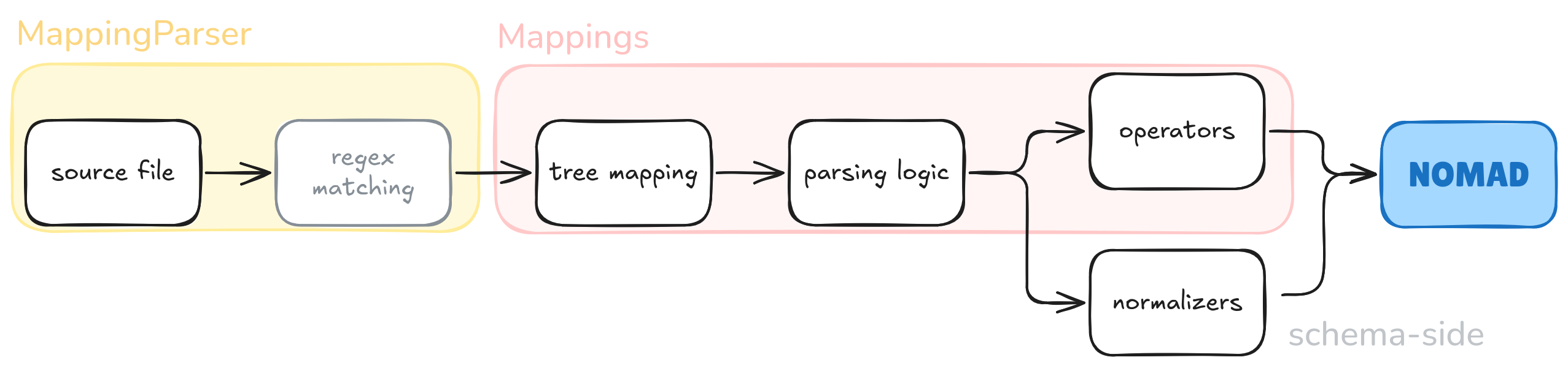
MappingParser is a NOMAD parsing framework for handling tree-like data structures.
It is extensively used for processing highly structured files, like computational output.
This framework acts as an intermediate step in the parsing phase, where reading the source file is done separtely from building up the archive (i.e. data).
Some functionality can be inserted in the mapping, the rest is relegated to normalization functionalitites of the schema.
Fundamentals¶
The NOMAD schema side targets several data paths within the source file.
These paths are represented using the JMesPath format.
This path is added to m_annotations, either overwriting or extending the previous annotations.
Each mapping corresponds to its own dictionary key, and a parser schema may contain multiple in parallel.
In the simplest case, e.g. a single quantity, you may simply write:
<quantity>.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(
out=MapperAnnotation(mapper=<OUTCAR jmes_path>),
xml=MapperAnnotation(mapper=<vasprun.xml jmes_path>),
)
)
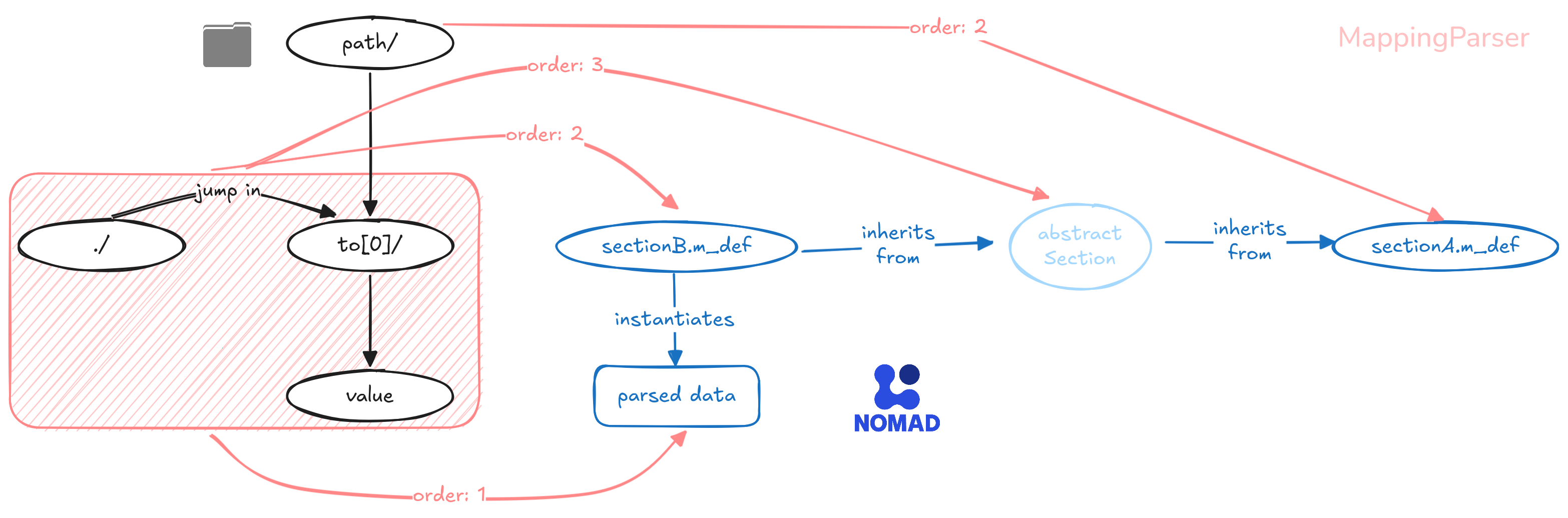
This works fine for top-level quantities, but those deeper down in the schema have to instantiate their containing sections.
These can be defined in absolute terms, or relative to their section above using the dot notation, e.g. .model.
The annotations themselves can be manipulated in two possible ways:
- definition level, i.e.
m_def. - attribute level, i.e. the data instance generated during runtime.
The mapping parser will search for the first available annotation in the following order:
- attribute level: specializes subsections to specific contexts.
- definition level: used for generic paths.
- child sections' definitions (i.e. all inheriting sections): used to bypass any abstract sections, which should not be instantiated themselves. (note)
In the case of quantities, there is no practical distinction between options 1 and 2. By convention, we use the shorter attribute-level annotation.
The following real-world example showcases the three annotation techniques listed above.
# GLOBAL m_def annotation
general.Simulation.m_def.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(xml=MapperAnnotation(mapper='modeling'))
)
# OPTIONAL CLASS INHERITANCE as a mask
class Simulation(general.Simulation): # Inherits from general.Simulation
-------------------------------------------------------------------------
2 POSSIBILITIES
# DEFINITION-LEVEL annotation (generic, default mapping)
general.Simulation.m_def.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(xml=MapperAnnotation(mapper='.parameters.separator'))
)
# ATTRIBUTE-LEVEL annotation (context-specific)
general.Simulation.program.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(xml=MapperAnnotation(mapper='.generator'))
)
-------------------------------------------------------------------------
# COMPOSE with the UPPER section's m_def (includes DFT as subsection)
model_method.DFT.m_def.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(xml=MapperAnnotation(mapper='.parameters.separator'))
)
# continue the mapping of the Program ATTRIBUTES
class Program(general.Program):
general.Program.version.m_annotations.setdefault(MAPPING_ANNOTATION_KEY, {}).update(
dict(xml=MapperAnnotation(mapper='.i[?"@name"==\'version\'] | [0].__value'))
)
Controlling Repeating Sections¶
Subsections that are defined as repeating in the schema are automatically picked up by the mapping parser. It will look for any repeating units along the path and instantiate the same number of subsections, i.e. \(\text{no. subsections} = \prod_{\text{segments}} \text{repeating path segment}\). To compose parallel branches into the same repeating subsection, i.e. \(\text{no. subsections} = \sum_{\text{branches}} \text{parallel branch}\):
- generate different mappings and add them to the annotation
dict. Each mapping comes with its own unique key. - during the conversion phase, select
update_mode="append". Manipulate the order in the data via the conversion order of the various maps.
Array quantities, meanwhile, with shape=['*'], have to be handled using an operator.
This is due to shape having variable rank, e.g. shape=['*','*'].
Other Edge Cases¶
Recursive sections, i.e. section schemas with the same schema as a subsection, should also be handled by an operator, or a different annotation key. Annotation keys that populate the same archive, can be loaded successively by the writer.
Likewise, references have to be set by the parser dev in the writer. They namely require a source and a target, and the schema only defines the source.
How-To¶
Use JMesPath¶
Accessing individual elements:
- Extract the first element:
Mapper(mapper='.array[0]') - Extract last element:
Mapper(mapper='.array[-1]') - Extract specific index with pipe syntax:
Mapper(mapper='.varray | [0].v')
Accessing multiple elements:
- Extract all elements:
Mapper(mapper='.array[*]') - Extract all vertex labels (exciting.py example):
Mapper(mapper=r'bandstructure.vertex[*]."@label"') - Extract from nested structure:
Mapper(mapper='.varray[?"@name"==\'kpointlist\'].v | [0]')
Conditional selection:
- Filter by attribute:
Mapper(mapper='.i[?"@name"==\'program\'] | [0].__value') - Filter by value comparison:
Mapper(mapper='c[?n>=1].v | [1]') - Complex filter with string matching:
Mapper(mapper='.separator[?"@name"==\'electronic exchange-correlation\']')
MappingParser¶
The MappingParser defines the source-to-archive mapping.
The source data_object can be an XML element tree or a metainfo section.
Settings include:
filepath: path to the input file to be parseddata_object: object resulting from loading the file in memory withload_filedata: dictionary representation ofdata_objectload_file: method to load the file given byfilepath
It also handles the actual conversion, via three methods.
Each requires a Mapper instance as input:
to_dict: method to convertdata_objectintodatafrom_dict: method to convertdataintodata_objectconvert: method to convert to another mapping parser
Conversion can be toggled via the update optional key.
| Update Mode | Description |
|---|---|
| replace | Replaces the target data entirely with the source data (default behavior) |
| append | Appends source data to target, preserving existing target values when they exist |
| merge | Merges dictionaries by recursively updating keys, or merges lists starting at index 0 |
| merge@start | Merges lists starting at the beginning (index 0) |
| merge@last | Merges lists starting from the end, calculated as len(source) - len(target) |
| merge@ |
Merges lists starting at a specific index n (can be negative for relative positioning) |
In the following, we describe the currently implemented mapping parsers.
XMLParser¶
This is mapping parser for XML files. It uses lxml to
load the file as an element tree. The dictionary is generated by iteratively parsing the
elements of the tree in to_dict. The values parsed from element text are automatically
converted to a corresponding data type. If attributes are present, the value is wrapped in
a dictionary with key given by value_key ('__value' by default) while the attribute keys
are prefixed by attribute_prefix ('@' by default). The following XML:
will be converted to:
data = {
'a' : {
'b': [
{'@name': 'item1', '__value': 'name'},
{'@name': 'item2', '__value': 'name2'}
]
}
}
The conversion can be reversed using the from_dict method.
HDF5Parser¶
This is the mapping parser for HDF5 files. It uses h5py to load
the file as an HDF5 group. Similar to XMLParser, the HDF5 datasets are
iteratively parsed from the underlying groups and if attributes are present these are
also parsed. The from_dict method is also implemented to convert a dictionary into an
HDF5 group.
MetainfoParser¶
This is the mapping parser for NOMAD archive files or metainfo sections.
It accepts a schema root node annotated with MappingAnnotation as data_object.
create_mapper generates the actual mapper as matching the annotation_key.
If a filepath is specified, it instead falls back on the ArchiveParser. (Note: Under development, more info on the ArchiveParser to be added).
The annotation should always point to a parsed value via a path (JMesPath format).
It may optionally specify a multi-argument operator for data mangling.
In this case, specify a tuple consisting of:
- the operator name, defined within the same scope.
- a list of paths with the corresponding values for the operator arguments.
Similar to MSection, it can be converted to (to_dict) or from (from_dict) a Python dict.
Other attributes are currently accessible.
from nomad.datamodel.metainfo.annotations import Mapper as MappingAnnotation
class BSection(ArchiveSection):
v = Quantity(type=np.float64, shape=[2, 2])
v.m_annotations['mapping'] = dict(
xml=MappingAnnotation(mapper='.v'),
hdf5=MappingAnnotation(mapper=('get_v', ['.v[0].d'])),
)
v2 = Quantity(type=str)
v2.m_annotations['mapping'] = dict(
xml=MappingAnnotation(mapper='.c[0].d[1]'),
hdf5=MappingAnnotation(mapper='g.v[-2]'),
)
class ExampleSection(ArchiveSection):
b = SubSection(sub_section=BSection, repeats=True)
b.m_annotations['mapping'] = dict(
xml=MappingAnnotation(mapper='a.b1'), hdf5=MappingAnnotation(mapper='.g1')
)
ExampleSection.m_def.m_annotations['mapping'] = dict(
xml=MappingAnnotation(mapper='a'), hdf5=MappingAnnotation(mapper='g')
)
parser = MetainfoParser()
p.data_object = ExampleSection(b=[BSection()])
p.annotation_key = 'xml'
p.mapper
# Mapper(source=Path(path='a'....
Converting mapping parsers¶
The following is a sample python code to illustrate the mapping of the contents of an
HDF5 file to an archive. First, we create a MetainfoParser object for the archive. The
annotation key is set to hdf5 which will generate a
mapper from the hdf5 annotations defined in the definitions. Essentially,
only metainfo sections and quantities with the hdf5 annotation will be mapped. The mapper
will contain paths for the source (HDF5) and the target (archive). The archive is then
set to the archive parser data_object. Here, the archive already contains some data
which should be merged to data that will be parsed. Next, a parser for HDF5 data is
created. We use a custom class of the HDF5Parser which implements the get_v method
defined in BSection.v In this example, we do not read the data from the HDF5 file but
instead generate it from a dictionary by using the from_dict method. By invoking the
convert method, the archive parser data object is populated with the corresponding
HDF5 data.
class ExampleHDF5Parser(HDF5Parser):
@staticmethod
def get_v(value):
return np.array(value)[1:, :2]
archive_parser = MetainfoParser()
archive_parser.annotation_key = 'hdf5'
archive_parser.data_object = ExampleSection(b=[BSection(v=np.eye(2))])
hdf5_parser = ExampleHDF5Parser()
d = dict(
g=dict(
g1=dict(v=[dict(d=np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]]))]),
v=['x', 'y', 'z'],
g=dict(
c1=dict(
i=[4, 6],
f=[
{'@index': 0, '__value': 1},
{'@index': 2, '__value': 2},
{'@index': 1, '__value': 1},
],
d=[dict(e=[3, 0, 4, 8, 1, 6]), dict(e=[1, 7, 8, 3, 9, 1])],
),
c=dict(v=[dict(d=np.eye(3), e=np.zeros(3)), dict(d=np.ones((3, 3)))]),
),
)
)
hdf5_parser.from_dict(d)
hdf5_parser.convert(archive_parser)
# >>> archive_parser.data_object
# ExampleSection(b, b2)
# >>> archive_parser.data_object.b[1].v
# array([[4., 5.],
# [7., 8.]])
Mapper¶
A mapper is necessary in order to convert a mapping parser to a target mapping parser
by mapping data from the source to the target. There are three kinds of mapper: Map,
Evaluate and Mapper each inheriting from BaseMapper. A mapper has attributes
source and target which define the paths to the source data and target, respectively.
Map is intended for mapping data directly from source to target. The path to the data is
given by the attribute path. Evaluate will execute a function defined by
function_name with the arguments given by the mapped values of the paths in
function_args. Lastly, Mapper allows the nesting of mappers by providing a list of
mappers to its attribute mapper. All the paths are instances of Path with the string
value of the path to the data given by the attribute path. The value of path should
follow the jmespath specifications but could be
prefixed by . which indicates that this is a path relative to the parent. This will communicate to the
mapper which source to get the data.
Mapper(
source=Path(path='a.b2', target=Path(path='b2'), mapper=[
Mapper(
source=Path(path='.c', parent=Path(path='a.b2')),
target=Path(path='.c', parent=Path(path='b2')), mapper=[
Map(
target=Path(
path='.i', parent=Path(path='.c', parent=Path(path='b2'))
),
path=Path(
path='.d', parent=Path(path='.c' parent=Path(path='a.b2'))
)
),
Evaluate(
target=Path(
path='.g', parent=Path(path='.c', parent=Path(path='b2'))
),
function_name='slice', function_args=[Path(path='a.b2.c.f.g.i')]
)
]
)
),
)