Python Reference¶
nomad.metainfo¶
nomad.config¶
This module describes all configurable parameters for the nomad python code. The configuration is used for all executed python code including API, worker, CLI, and other scripts. To use the configuration in your own scripts or new modules, simply import this module.
All parameters are structured into objects for two reasons. First, to have
categories. Second, to allow runtime manipulation that is not effected
by python import logic. The categories are choosen along infrastructure components:
mongo, elastic, etc.
This module also provides utilities to read the configuration from environment variables and .yaml files. This is done automatically on import. The precedence is env over .yaml over defaults.
-
class
nomad.config.NomadConfig(**kwargs)¶ A class for configuration categories. It is a dict subclass that uses attributes as key/value pairs.
-
__init__(**kwargs)¶ Initialize self. See help(type(self)) for accurate signature.
-
-
nomad.config.load_config()¶ Loads the configuration from nomad.yaml and environment.
nomad.infrastructure¶
This module provides function to establish connections to the database, searchengine, etc.
infrastructure services. Usually everything is setup at once with setup(). This
is run once for each api and worker process. Individual functions for partial setups
exist to facilitate testing, aspects of nomad.cli, etc.
-
nomad.infrastructure.elastic_client= None¶ The elastic search client.
-
nomad.infrastructure.mongo_client= None¶ The pymongo mongodb client.
-
nomad.infrastructure.setup()¶ Uses the current configuration (nomad/config.py and environment) to setup all the infrastructure services (repository db, mongo, elastic search) and logging. Will create client instances for the databases and has to be called before they can be used.
-
nomad.infrastructure.setup_files()¶
-
nomad.infrastructure.setup_mongo(client=False)¶ Creates connection to mongodb.
-
nomad.infrastructure.setup_elastic(create_mappings=True)¶ Creates connection to elastic search.
-
exception
nomad.infrastructure.KeycloakError¶
-
class
nomad.infrastructure.Keycloak¶ A class that encapsulates all keycloak related functions for easier mocking and configuration
-
__init__()¶ Initialize self. See help(type(self)) for accurate signature.
-
auth(headers: Dict[str, str], allow_basic: bool = False) → Tuple[object, str]¶ Performs authentication based on the provided headers. Either basic or bearer.
- Returns
The user and its access_token
- Raises
-
basicauth(username: str, password: str) → str¶ Performs basic authentication and returns an access token.
- Raises
-
tokenauth(access_token: str) → object¶ Authenticates the given access_token
- Returns
The user
- Raises
-
add_user(user, bcrypt_password=None, invite=False)¶ Adds the given
nomad.datamodel.Userinstance to the configured keycloak realm using the keycloak admin API.
-
search_user(query: str = None, max=1000, **kwargs)¶
-
get_user(user_id: str = None, username: str = None, user=None) → object¶ Retrives all available information about a user from the keycloak admin interface. This must be used to retrieve complete user information, because the info solely gathered from tokens is generally incomplete.
-
-
nomad.infrastructure.reset(remove: bool)¶ Resets the databases mongo, elastic/calcs, and all files. Be careful. In contrast to
remove(), it will only remove the contents of dbs and indicies. This function just attempts to remove everything, there is no exception handling or any warranty it will succeed.- Parameters
remove – Do not try to recreate empty databases, remove entirely.
-
nomad.infrastructure.send_mail(name: str, email: str, message: str, subject: str)¶ Used to programmatically send mails.
- Parameters
name – The email recipient name.
email – The email recipient address.
messsage – The email body.
subject – The subject line.
nomad.datamodel¶
Introduction¶
This is about the datamodel that is used to represent NOMAD entries in our databases and search engines. The respective data, also known as (repository) metadata is also part of the NOMAD Archive and the datamodel is also defined based on the NOMAD Metainfo (section metadata). It covers all information that users can search for and that can be easily rendered on the GUI. The information is readily available through the repo API.
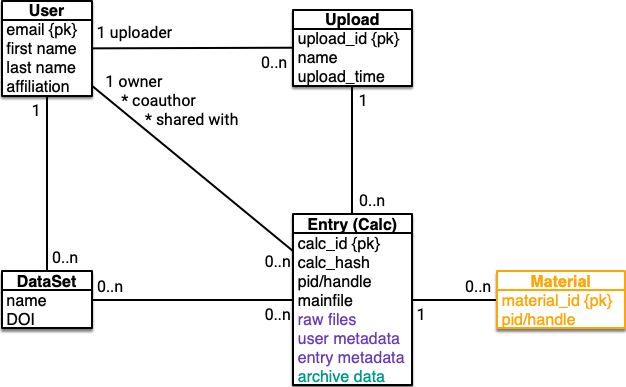
See also the datamodel section in the introduction.
This module contains classes that allow to represent the core
nomad data entities (entries/calculations, users, datasets) on a high level of abstraction
independent from their representation in the different modules
nomad.processing, nomad.parsing, nomad.search, nomad.app.
Datamodel entities¶
The entities in the datamodel are defined as NOMAD Metainfo sections. They are treated similarily to all Archive data. The entry/calculation datamodel data is created during processing. It is not about representing every detail, but those parts that are directly involved in api, processing, mirroring, or other ‘infrastructure’ operations.
The class User is used to represent users and their attributes.
-
class
nomad.datamodel.User(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ A NOMAD user.
Typically a NOMAD user has a NOMAD account. The user related data is managed by NOMAD keycloak user-management system. Users are used to denote uploaders, authors, people to shared data with embargo with, and owners of datasets.
- Parameters
user_id – The unique, persistent keycloak UUID
username – The unique, persistent, user chosen username
first_name – The users first name (including all other given names)
last_name – The users last name
affiliation – The name of the company and institutes the user identifies with
affiliation_address – The address of the given affiliation
create – The time the account was created
repo_user_id – The id that was used to identify this user in the NOMAD CoE Repository
is_admin – Bool that indicated, iff the user the use admin user
-
m_def: Section = nomad.datamodel.datamodel.User:Section¶
-
user_id¶ An optimized replacement for Quantity suitable for primitive properties.
-
username¶ An optimized replacement for Quantity suitable for primitive properties.
-
created¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type¶ Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape¶ The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar¶ Derived quantity that is True, iff this quantity has shape of length 0
-
unit¶ The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default¶ The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived¶ A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached¶ A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual¶ A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
repo_user_id¶ Optional, legacy user id from the old NOMAD CoE repository.
-
is_admin¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar Derived quantity that is True, iff this quantity has shape of length 0
-
unit The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
is_oasis_admin¶ An optimized replacement for Quantity suitable for primitive properties.
-
static
get(*args, **kwargs) → nomad.datamodel.datamodel.User¶
-
full_user() → nomad.datamodel.datamodel.User¶ Returns a User object with all attributes loaded from the user management system.
The class Dataset is used to represent datasets and their attributes.
-
class
nomad.datamodel.Dataset(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ A Dataset is attached to one or many entries to form a set of data.
- Parameters
dataset_id – The unique identifier for this dataset as a string. It should be a randomly generated UUID, similar to other nomad ids.
name – The human readable name of the dataset as string. The dataset name must be unique for the user.
user_id – The unique user_id of the owner and creator of this dataset. The owner must not change after creation.
doi – The optional Document Object Identifier (DOI) associated with this dataset. Nomad can register DOIs that link back to the respective representation of the dataset in the nomad UI. This quantity holds the string representation of this DOI. There is only one per dataset. The DOI is just the DOI name, not its full URL, e.g. “10.17172/nomad/2019.10.29-1”.
pid – The original NOMAD CoE Repository dataset PID. Old DOIs still reference datasets based on this id. Is not used for new datasets.
created – The date when the dataset was first created.
modified – The date when the dataset was last modified. An owned dataset can only be extended after a DOI was assigned. A foreign dataset cannot be changed once a DOI was assigned.
dataset_type – The type determined if a dataset is owned, i.e. was created by the uploader/owner of the contained entries; or if a dataset is foreign, i.e. it was created by someone not necessarily related to the entries.
-
m_def: Section = nomad.datamodel.datamodel.Dataset:Section¶
-
dataset_id¶ An optimized replacement for Quantity suitable for primitive properties.
-
name¶ An optimized replacement for Quantity suitable for primitive properties.
-
user_id¶ An optimized replacement for Quantity suitable for primitive properties.
-
doi¶ An optimized replacement for Quantity suitable for primitive properties.
-
pid¶ An optimized replacement for Quantity suitable for primitive properties.
-
created¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type¶ Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape¶ The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar¶ Derived quantity that is True, iff this quantity has shape of length 0
-
unit¶ The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default¶ The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived¶ A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached¶ A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual¶ A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
modified¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar Derived quantity that is True, iff this quantity has shape of length 0
-
unit The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
dataset_type¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar Derived quantity that is True, iff this quantity has shape of length 0
-
unit The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
query¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar Derived quantity that is True, iff this quantity has shape of length 0
-
unit The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
entries¶ An optimized replacement for Quantity suitable for primitive properties.
The class MongoMetadata is used to tag metadata stored in mongodb.
-
class
nomad.datamodel.MongoMetadata¶ NOMAD entry quantities that are stored in mongodb and not necessarely in the archive.
-
m_def: Category = nomad.datamodel.datamodel.MongoMetadata:Category¶
-
The class EntryMetadata is used to represent all metadata about an entry.
-
class
nomad.datamodel.EntryMetadata(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
upload_id¶ The
upload_idof the calculations upload (random UUID).
-
calc_id¶ The unique mainfile based calculation id.
-
calc_hash¶ The raw file content based checksum/hash of this calculation.
-
pid¶ The unique persistent id of this calculation.
-
mainfile¶ The upload relative mainfile path.
-
domain¶ Must be the key for a registered domain. This determines which actual subclass is instantiated.
-
files¶ A list of all files, relative to upload.
-
upload_time¶ The time when the calc was uploaded.
-
uploader¶ An object describing the uploading user, has at least
user_id
-
processed¶ Boolean indicating if this calc was successfully processed and archive data and calc metadata is available.
-
last_processing¶ A datatime with the time of the last successful processing.
-
nomad_version¶ A string that describes the version of the nomad software that was used to do the last successful processing.
-
comment¶ An arbitrary string with user provided information about the entry.
-
references¶ A list of URLs for resources that are related to the entry.
-
uploader Id of the uploader of this entry.
Ids of all co-authors (excl. the uploader) of this entry. Co-authors are shown as authors of this entry alongside its uploader.
Ids of all users that this entry is shared with. These users can find, see, and download all data for this entry, even if it is in staging or has an embargo.
-
with_embargo¶ Entries with embargo are only visible to the uploader, the admin user, and users the entry is shared with (see shared_with).
-
upload_time The time that this entry was uploaded
-
datasets¶ Ids of all datasets that this entry appears in
-
m_def: Section = nomad.datamodel.datamodel.EntryMetadata:Section¶
-
upload_id The persistent and globally unique identifier for the upload of the entry
-
calc_id A persistent and globally unique identifier for the entry
-
calc_hash A raw file content based checksum/hash
-
mainfile The path to the mainfile from the root directory of the uploaded files
-
files The paths to the files within the upload that belong to this entry. All files within the same directory as the entry’s mainfile are considered the auxiliary files that belong to the entry.
-
pid The unique, sequentially enumerated, integer PID that was used in the legacy NOMAD CoE. It allows to resolve URLs of the old NOMAD CoE Repository.
-
raw_id¶ The code specific identifier extracted from the entrie’s raw files if such an identifier is supported by the underlying code
-
domain The material science domain
-
published¶ Indicates if the entry is published
-
processed Indicates that the entry is successfully processed.
-
last_processing The datetime of the last processing
-
processing_errors¶ Errors that occured during processing
-
nomad_version The NOMAD version used for the last processing
-
nomad_commit¶ The NOMAD commit used for the last processing
-
parser_name¶ The NOMAD parser used for the last processing
-
comment A user provided comment for this entry
-
references User provided references (URLs) for this entry
-
external_db¶ The repository or external database where the original data resides
-
uploader The uploader of the entry
-
origin¶ A short human readable description of the entries origin. Usually it is the handle of an external database/repository or the name of the uploader.
-
coauthors A user provided list of co-authors
All authors (uploader and co-authors)
-
shared_with A user provided list of userts to share the entry with
-
owners¶ All owner (uploader and shared with users)
-
license¶ A short license description (e.g. CC BY 4.0), that refers to the license of this entry.
-
with_embargo Indicated if this entry is under an embargo
-
upload_time The date and time this entry was uploaded to nomad
-
upload_name¶ The user provided upload name
-
datasets A list of user curated datasets this entry belongs to.
-
external_id¶ A user provided external id. Usually the id for an entry in an external database where the data was imported from.
-
last_edit¶ The date and time the user metadata was edited last
-
formula¶ A (reduced) chemical formula
-
atoms¶ The atom labels of all atoms of the entry’s material
-
only_atoms¶ The atom labels concatenated in order-number order
-
n_atoms¶ The number of atoms in the entry’s material
-
ems¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
dft¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
qcms¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
encyclopedia¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
apply_user_metadata(metadata: dict)¶ Applies a user provided metadata dict to this calc.
-
apply_domain_metadata(archive)¶ Used to apply metadata that is related to the domain.
-
Domains¶
The datamodel supports different domains. This means that most domain metadata of an
entry/calculation is stored in domain-specific sub sections of the EntryMetadata
section. We currently have the following domain specific metadata classes/sections:
-
class
nomad.datamodel.dft.DFTMetadata(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.dft.DFTMetadata:Section¶
-
basis_set¶ The used basis set functions.
-
xc_functional¶ The libXC based xc functional classification used in the simulation.
-
xc_functional_names¶ The list of libXC functional names that where used in this entry.
-
system¶ The system type of the simulated system.
-
compound_type¶ The compound type of the simulated system.
-
crystal_system¶ The crystal system type of the simulated system.
-
spacegroup¶ The spacegroup of the simulated system as number.
-
spacegroup_symbol¶ The spacegroup as international short symbol.
-
code_name¶ The name of the used code.
-
code_version¶ The version of the used code.
-
n_geometries¶ Number of unique geometries.
-
n_calculations¶ Number of single configuration calculation sections
-
n_total_energies¶ Number of total energy calculations
-
n_quantities¶ Number of metainfo quantities parsed from the entry.
-
quantities¶ All quantities that are used by this entry.
-
searchable_quantities¶ All quantities with existence filters in the search GUI.
-
geometries¶ Hashes for each simulated geometry
-
group_hash¶ Hashes that describe unique geometries simulated by this code run.
-
labels¶ The labels taken from AFLOW prototypes and springer.
-
labels_springer_compound_class¶ Springer compund classification.
-
labels_springer_classification¶ Springer classification by property.
-
optimade¶ Metadata used for the optimade API.
-
workflow¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type¶ Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape¶ The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar¶ Derived quantity that is True, iff this quantity has shape of length 0
-
unit¶ The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default¶ The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived¶ A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached¶ A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual¶ A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
code_name_from_parser()¶
-
update_group_hash()¶
-
apply_domain_metadata(entry_archive)¶
-
band_structure_electronic(entry_archive)¶ - Returns whether a valid electronic band structure can be found. In
the case of multiple valid band structures, only the latest one is considered.
- Band structure is reported only under the following conditions:
There is a non-empty array of band_k_points.
There is a non-empty array of band_energies.
The reported band_structure_kind is not “vibrational”.
-
dos_electronic(entry_archive)¶ - Returns whether a valid electronic DOS can be found. In the case of
multiple valid DOSes, only the latest one is reported.
- DOS is reported only under the following conditions:
There is a non-empty array of dos_values_normalized.
There is a non-empty array of dos_energies.
The reported dos_kind is not “vibrational”.
-
band_structure_phonon(entry_archive)¶ - Returns whether a valid phonon band structure can be found. In the
case of multiple valid band structures, only the latest one is considered.
- Band structure is reported only under the following conditions:
There is a non-empty array of band_k_points.
There is a non-empty array of band_energies.
The reported band_structure_kind is “vibrational”.
-
dos_phonon(entry_archive)¶ - Returns whether a valid phonon dos can be found. In the case of
multiple valid data sources, only the latest one is reported.
- DOS is reported only under the following conditions:
There is a non-empty array of dos_values_normalized.
There is a non-empty array of dos_energies.
The reported dos_kind is “vibrational”.
-
traverse_reversed(entry_archive, path)¶ Traverses the given metainfo path in reverse order. Useful in finding the latest reported section or value.
-
-
class
nomad.datamodel.ems.EMSMetadata(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.ems.EMSMetadata:Section¶
-
chemical¶ An optimized replacement for Quantity suitable for primitive properties.
-
sample_constituents¶ An optimized replacement for Quantity suitable for primitive properties.
-
sample_microstructure¶ An optimized replacement for Quantity suitable for primitive properties.
-
experiment_summary¶ An optimized replacement for Quantity suitable for primitive properties.
-
origin_time¶ To define quantities, instantiate
Quantityas a classattribute values in a section classes. The name of a quantity is automatically taken from its section class attribute. You can provide all other attributes to the constructor with keyword argumentsSee metainfo-sections to learn about section classes. In Python terms,
Quantityis a descriptor. Descriptors define how to get and set attributes in a Python object. This allows us to use sections like regular Python objects and quantity like regular Python attributes.Each quantity must define a basic data type and a shape. The values of a quantity must fulfil the given type. The default shape is a single value. Quantities can also have physical units. Units are applied to all values.
-
type¶ Defines the datatype of quantity values. This is the type of individual elements in a potentially complex shape. If you define a list of integers for example, the shape would be list and the type integer:
Quantity(type=int, shape=['0..*']).The type can be one of:
a build-in primitive Python type:
int,str,bool,floatan instance of
MEnum, e.g.MEnum('one', 'two', 'three')a section to define references to other sections as quantity values
a custom meta-info
DataType, see Environmentsa numpy dtype, e.g.
np.dtype('float32')typing.Anyto support any value
If set to dtype, this quantity will use a numpy array or scalar to store values internally. If a regular (nested) Python list or Python scalar is given, it will be automatically converted. The given dtype will be used in the numpy value.
To define a reference, either a section class or instance of
Sectioncan be given. See metainfo-sections for details. Instances of the given section constitute valid values for this type. Upon serialization, references section instance will represented with metainfo URLs. See Resources.For quantities with more than one dimension, only numpy arrays and dtypes are allowed.
-
shape¶ The shape of the quantity. It defines its dimensionality.
A shape is a list, where each item defines one dimension. Each dimension can be:
an integer that defines the exact size of the dimension, e.g.
[3]is the shape of a 3D spacial vectora string that specifies a possible range, e.g.
0..*,1..*,3..6the name of an int typed and shapeless quantity in the same section which values define the length of this dimension, e.g.
number_of_atomsdefines the length ofatom_positions
Range specifications define lower and upper bounds for the possible dimension length. The
*can be used to denote an arbitrarily high upper bound.Quantities with dimensionality (length of the shape) higher than 1, must be numpy arrays. Theire type must be a dtype.
-
is_scalar¶ Derived quantity that is True, iff this quantity has shape of length 0
-
unit¶ The physics unit for this quantity. It is optional.
Units are represented with the Pint Python package. Pint defines units and their algebra. You can either use pint units directly, e.g.
units.m / units.s. The metainfo provides a preconfigured pint unit registryureg. You can also provide the unit as pint parsable string, e.g.'meter / seconds'or'm/s'.
-
default¶ The default value for this quantity. The value must match type and shape.
Be careful with a default value like
[]as it will be the default value for all occurrences of this quantity.
Quantities are mapped to Python properties on all section objects that instantiate the Python class/section definition that has this quantity. This means quantity values can be read and set like normal Python attributes.
In some cases it might be desirable to have virtual and read only quantities that are not real quantities used for storing values, but rather define an interface to other quantities. Examples for this are synonyms and derived quantities.
-
derived¶ A Python callable that takes the containing section as input and outputs the value for this quantity. This quantity cannot be set directly, its value is only derived by the given callable. The callable is executed when this quantity is get. Derived quantities are always virtual.
-
cached¶ A bool indicating that derived values should be cached unless the underlying section has changed.
-
virtual¶ A boolean that determines if this quantity is virtual. Virtual quantities can be get/set like regular quantities, but their values are not (de-)serialized, hence never permanently stored.
-
-
experiment_location¶ An optimized replacement for Quantity suitable for primitive properties.
-
method¶ An optimized replacement for Quantity suitable for primitive properties.
-
data_type¶ An optimized replacement for Quantity suitable for primitive properties.
-
probing_method¶ An optimized replacement for Quantity suitable for primitive properties.
-
repository_name¶ An optimized replacement for Quantity suitable for primitive properties.
-
repository_url¶ An optimized replacement for Quantity suitable for primitive properties.
-
entry_repository_url¶ An optimized replacement for Quantity suitable for primitive properties.
-
preview_url¶ An optimized replacement for Quantity suitable for primitive properties.
-
quantities¶ An optimized replacement for Quantity suitable for primitive properties.
-
group_hash¶ An optimized replacement for Quantity suitable for primitive properties.
-
apply_domain_metadata(entry_archive)¶
-
-
class
nomad.datamodel.OptimadeEntry(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.optimade.OptimadeEntry:Section¶
-
elements¶ Names of the different elements present in the structure.
-
nelements¶ Number of different elements in the structure as an integer.
-
elements_ratios¶ Relative proportions of different elements in the structure.
-
chemical_formula_descriptive¶ The chemical formula for a structure as a string in a form chosen by the API implementation.
-
chemical_formula_reduced¶ The reduced chemical formula for a structure as a string with element symbols and integer chemical proportion numbers. The proportion number MUST be omitted if it is 1.
-
chemical_formula_hill¶ The chemical formula for a structure in Hill form with element symbols followed by integer chemical proportion numbers. The proportion number MUST be omitted if it is 1.
-
chemical_formula_anonymous¶ The anonymous formula is the chemical_formula_reduced, but where the elements are instead first ordered by their chemical proportion number, and then, in order left to right, replaced by anonymous symbols A, B, C, …, Z, Aa, Ba, …, Za, Ab, Bb, … and so on.
-
dimension_types¶ List of three integers. For each of the three directions indicated by the three lattice vectors (see property lattice_vectors). This list indicates if the direction is periodic (value 1) or non-periodic (value 0). Note: the elements in this list each refer to the direction of the corresponding entry in lattice_vectors and not the Cartesian x, y, z directions.
-
nperiodic_dimensions¶ An integer specifying the number of periodic dimensions in the structure, equivalent to the number of non-zero entries in dimension_types.
-
lattice_vectors¶ The three lattice vectors in Cartesian coordinates, in ångström (Å).
-
cartesian_site_positions¶ Cartesian positions of each site. A site is an atom, a site potentially occupied by an atom, or a placeholder for a virtual mixture of atoms (e.g., in a virtual crystal approximation).
-
nsites¶ An integer specifying the length of the cartesian_site_positions property.
-
species_at_sites¶ Name of the species at each site (where values for sites are specified with the same order of the cartesian_site_positions property). The properties of the species are found in the species property.
-
structure_features¶ A list of strings that flag which special features are used by the structure.
disorder: This flag MUST be present if any one entry in the species list has a
chemical_symbols list that is longer than 1 element. - unknown_positions: This flag MUST be present if at least one component of the cartesian_site_positions list of lists has value null. - assemblies: This flag MUST be present if the assemblies list is present.
-
species¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
-
class
nomad.datamodel.encyclopedia.WyckoffVariables(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.WyckoffVariables:Section¶
-
x¶ The x variable if present.
-
y¶ The y variable if present.
-
z¶ The z variable if present.
-
-
class
nomad.datamodel.encyclopedia.WyckoffSet(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.WyckoffSet:Section¶
-
wyckoff_letter¶ The Wyckoff letter for this set.
-
indices¶ Indices of the atoms belonging to this group.
-
element¶ Chemical element at this Wyckoff position.
-
variables¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
-
class
nomad.datamodel.encyclopedia.LatticeParameters(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.LatticeParameters:Section¶
-
a¶ Length of the first basis vector.
-
b¶ Length of the second basis vector.
-
c¶ Length of the third basis vector.
-
alpha¶ Angle between second and third basis vector.
-
beta¶ Angle between first and third basis vector.
-
gamma¶ Angle between first and second basis vector.
-
-
class
nomad.datamodel.encyclopedia.IdealizedStructure(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.IdealizedStructure:Section¶
-
atom_labels¶ Type (element, species) of each atom.
-
atom_positions¶ Atom positions given in coordinates that are relative to the idealized cell.
-
lattice_vectors¶ Lattice vectors of the idealized structure. For bulk materials it is the Bravais cell. This cell is representative and is idealized to match the detected symmetry properties.
-
lattice_vectors_primitive¶ Lattice vectors of the the primitive unit cell in a form to be visualized within the idealized cell. This cell is representative and is idealized to match the detected symmemtry properties.
-
periodicity¶ Automatically detected true periodicity of each lattice direction. May not correspond to the periodicity used in the calculation.
-
number_of_atoms¶ Number of atoms in the idealized structure.”
-
cell_volume¶ Volume of the idealized cell. The cell volume can only be reported consistently after idealization and may not perfectly correspond to the original simulation cell.
-
wyckoff_sets¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
lattice_parameters¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
-
class
nomad.datamodel.encyclopedia.Bulk(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Bulk:Section¶
-
bravais_lattice¶ The Bravais lattice type in the Pearson notation, where the first lowercase letter indicates the crystal system, and the second uppercase letter indicates the lattice type. The value can only be one of the 14 different Bravais lattices in three dimensions.
Crystal system letters:
a = Triclinic m = Monoclinic o = Orthorhombic t = Tetragonal h = Hexagonal and Trigonal c = Cubic
Lattice type letters:
P = Primitive S (A, B, C) = One side/face centred I = Body centered R = Rhombohedral centring F = All faces centred
-
crystal_system¶ The detected crystal system. One of seven possibilities in three dimensions.
-
has_free_wyckoff_parameters¶ Whether the material has any Wyckoff sites with free parameters. If a materials has free Wyckoff parameters, at least some of the atoms are not bound to a particular location in the structure but are allowed to move with possible restrictions set by the symmetry.
-
point_group¶ Point group in Hermann-Mauguin notation, part of crystal structure classification. There are 32 point groups in three dimensional space.
-
space_group_number¶ Integer representation of the space group, part of crystal structure classification, part of material definition.
-
space_group_international_short_symbol¶ International short symbol notation of the space group.
-
structure_prototype¶ The prototypical material for this crystal structure.
-
structure_type¶ Classification according to known structure type, considering the point group of the crystal and the occupations with different atom types.
-
strukturbericht_designation¶ Classification of the material according to the historically grown “strukturbericht”.
-
-
class
nomad.datamodel.encyclopedia.Material(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Material:Section¶
-
material_type¶ “Broad structural classification for the material, e.g. bulk, 2D, 1D… “,
-
material_id¶ A fixed length, unique material identifier in the form of a hash digest.
-
material_name¶ Most meaningful name for a material if one could be assigned
-
material_classification¶ Contains the compound class and classification of the material according to springer materials in JSON format.
-
formula¶ Formula giving the composition and occurrences of the elements in the Hill notation. For periodic materials the formula is calculated fom the primitive unit cell.
-
formula_reduced¶ Formula giving the composition and occurrences of the elements in the Hill notation where the number of occurences have been divided by the greatest common divisor.
-
species_and_counts¶ The formula separated into individual terms containing both the atom type and count. Used for searching parts of a formula.
-
species¶ The formula separated into individual terms containing only unique atom species. Used for searching materials containing specific elements.
-
bulk¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
idealized_structure¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
-
class
nomad.datamodel.encyclopedia.Method(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Method:Section¶
-
method_type¶ Generic name for the used methodology.
-
core_electron_treatment¶ How the core electrons are described.
-
functional_long_name¶ Full identified for the used exchange-correlation functional.
-
functional_type¶ Basic type of the used exchange-correlation functional.
-
method_id¶ A fixed length, unique method identifier in the form of a hash digest. The hash is created by using several method settings as seed. This hash is only defined if a set of well-defined method settings is available for the used program.
-
group_eos_id¶ A fixed length, unique identifier for equation-of-state calculations. Only calculations within the same upload and with a method hash available will be grouped under the same hash.
-
group_parametervariation_id¶ A fixed length, unique identifier for calculations where structure is identical but the used computational parameters are varied. Only calculations within the same upload and with a method hash available will be grouped under the same hash.
-
gw_starting_point¶ The exchange-correlation functional that was used as a starting point for this GW calculation.
-
gw_type¶ Basic type of GW calculation.
-
smearing_kind¶ Smearing function used for the electronic structure calculation.
-
smearing_parameter¶ Parameter for smearing, usually the width.
-
-
class
nomad.datamodel.encyclopedia.Calculation(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Calculation:Section¶
-
calculation_type¶ Defines the type of calculation that was detected for this entry.
-
-
class
nomad.datamodel.encyclopedia.Energies(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Energies:Section¶
-
energy_total¶ Total energy.
-
energy_total_T0¶ Total energy projected to T=0.
-
energy_free¶ Free energy.
-
-
class
nomad.datamodel.encyclopedia.Properties(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.Properties:Section¶
-
atomic_density¶ Atomic density of the material (atoms/volume).”
-
mass_density¶ Mass density of the material.
-
band_gap¶ Band gap value. If multiple spin channels are present, this value is taken from the channel with smallest band gap value.
-
band_gap_direct¶ Whether band gap is direct or not. If multiple spin channels are present, this value is taken from the channel with smallest band gap value.
-
energies¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
electronic_band_structure¶ Reference to an electronic band structure.
-
electronic_dos¶ Reference to an electronic density of states.
-
phonon_band_structure¶ Reference to a phonon band structure.
-
phonon_dos¶ Reference to a phonon density of states.
-
thermodynamical_properties¶ Reference to a section containing thermodynamical properties.
-
-
class
nomad.datamodel.encyclopedia.EncyclopediaMetadata(m_def: Optional[nomad.metainfo.metainfo.Section] = None, m_resource: nomad.metainfo.metainfo.MResource = None, **kwargs)¶ -
m_def: Section = nomad.datamodel.encyclopedia.EncyclopediaMetadata:Section¶
-
material¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section¶ A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats¶ A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
method¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
properties¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
calculation¶ Like quantities, sub-sections are defined in a section class as attributes of this class. An like quantities, each sub-section definition becomes a property of the corresponding section definition (parent). A sub-section definition references another section definition as the sub-section (child). As a consequence, parent section instances can contain child section instances as sub-sections.
Contrary to the old NOMAD metainfo, we distinguish between sub-section the section and sub-section the property. This allows to use on child section definition as sub-section of many different parent section definitions.
-
sub_section A
Sectionor Python class object for a section class. This will be the child section definition. The defining section the child section definition.
-
repeats A boolean that determines wether this sub-section can appear multiple times in the parent section.
-
-
status¶ The final Encyclopedia processing status for this entry. The meaning of the status is as follows:
Status | Description |——————————– | —————————————————————————– |“success” | Processed successfully |“unsupported_material_type” | The detected material type is currently not supported by the Encyclopedia. |“unsupported_calculation_type” | The detected calculation type is currently not supported by the Encyclopedia. |“invalid_metainfo” | The entry could not be processed due to missing or invalid metainfo. |“failure” | The entry could not be processed due to an unexpected exception. |
-
nomad.files¶
Uploads contains classes and functions to create and maintain file structures for uploads.
There are two different structures for uploads in two different states: staging and public. Possible operations on uploads differ based on this state. Staging is used for processing, heavily editing, creating hashes, etc. Public is supposed to be a almost readonly (beside metadata) storage.
fs/staging/<upload>/raw/**
/archive/<calc>.json
fs/public/<upload>/raw-public.plain.zip
/raw-restricted.plain.zip
/archive-public.json.zip
/archive-restricted.json.zip
There is an implicit relationship between files, based on them being in the same directory. Each directory with at least one mainfile is a calculation directory and all the files are aux files to that mainfile. This is independent of the respective files actually contributing data or not. A calculation directory might contain multiple mainfile. E.g., user simulated multiple states of the same system, have one calculation based on the other, etc. In this case the other mainfile is an aux file to the original mainfile and vice versa.
Published files are kept in pairs of public and restricted files. Here the multiple mainfiles per directory provides a dilemma. If on mainfile is restricted, all its aux files should be restricted too. But if one of the aux files is actually a mainfile it might be published!
There are multiple ways to solve this. Due to the rarity of the case, we take the most simple solution: if one file is public, all files are made public, execpt those being other mainfiles. Therefore, the aux files of a restricted calc might become public!
-
nomad.files.always_restricted(path: str)¶ Used to put general restrictions on files, e.g. due to licensing issues. Will be called during packing and while accessing public files.
-
nomad.files.copytree(src, dst)¶ A close on
shutils.copytreethat does not try to copy the stats on all files. This is unecessary for our usecase and also causes permission denies for unknown reasons.
-
class
nomad.files.PathObject(bucket: str, object_id: str, os_path: str = None, prefix: bool = False, create_prefix: bool = False)¶ Object storage-like abstraction for paths in general. :param bucket: The bucket to store this object in :param object_id: The object id (i.e. directory path) :param os_path: Override the “object storage” path with the given path. :param prefix: Add a x-digit prefix directory, e.g. foo/test/ -> foo/tes/test :param create_prefix: Create the prefix right away
-
__init__(bucket: str, object_id: str, os_path: str = None, prefix: bool = False, create_prefix: bool = False) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
delete() → None¶
-
exists() → bool¶
-
property
size¶ The os determined file size.
-
-
class
nomad.files.DirectoryObject(bucket: str, object_id: str, create: bool = False, **kwargs)¶ Object storage-like abstraction for directories. :param bucket: The bucket to store this object in :param object_id: The object id (i.e. directory path) :param create: True if the directory structure should be created. Default is False.
-
__init__(bucket: str, object_id: str, create: bool = False, **kwargs) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
join_dir(path, create: bool = None) → nomad.files.DirectoryObject¶
-
join_file(path) → nomad.files.PathObject¶
-
exists() → bool¶
-
-
exception
nomad.files.ExtractError¶
-
exception
nomad.files.Restricted¶
-
class
nomad.files.UploadFiles(bucket: str, upload_id: str, is_authorized: Callable[[], bool] = <function UploadFiles.<lambda>>, create: bool = False)¶ -
__init__(bucket: str, upload_id: str, is_authorized: Callable[[], bool] = <function UploadFiles.<lambda>>, create: bool = False) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
property
user_metadata¶
-
to_staging_upload_files(create: bool = False, **kwargs) → nomad.files.StagingUploadFiles¶ Casts to or creates corresponding staging upload files or returns None.
-
static
get(upload_id: str, *args, **kwargs) → nomad.files.UploadFiles¶
-
raw_file(file_path: str, *args, **kwargs) → IO¶ Opens a raw file and returns a file-like object. Additional args, kwargs are delegated to the respective open call. :param file_path: The path to the file relative to the upload.
- Raises
KeyError – If the file does not exist.
Restricted – If the file is restricted and upload access evaluated to False.
-
raw_file_size(file_path: str) → int¶ - Returns
The size of the given raw file.
-
raw_file_manifest(path_prefix: str = None) → Generator[str, None, None]¶ Returns the path for all raw files in the archive (with a given prefix). :param path_prefix: An optional prefix; only returns those files that have the prefix.
- Returns
An iterable over all (matching) raw files.
-
raw_file_list(directory: str) → List[Tuple[str, int]]¶ Gives a list of directory contents and its size. :param directory: The directory to list
- Returns
A list of tuples with file name and size.
-
read_archive(calc_id: str, access: str = None) → nomad.archive.storage.ArchiveReader¶ Returns an
nomad.archive.ArchiveReaderthat contains the given calc_id. Both restricted and public archive are searched by default. The optionalaccessparameter can be used to limit this lookup to thepublicorrestrictedarchive.
-
close()¶ Release possibly held system resources (e.g. file handles).
-
-
class
nomad.files.StagingUploadFiles(upload_id: str, is_authorized: Callable[[], bool] = <function StagingUploadFiles.<lambda>>, create: bool = False)¶ -
__init__(upload_id: str, is_authorized: Callable[[], bool] = <function StagingUploadFiles.<lambda>>, create: bool = False) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
to_staging_upload_files(create: bool = False, **kwargs) → nomad.files.StagingUploadFiles¶ Casts to or creates corresponding staging upload files or returns None.
-
property
size¶ The os determined file size.
-
raw_file(file_path: str, *args, **kwargs) → IO¶ Opens a raw file and returns a file-like object. Additional args, kwargs are delegated to the respective open call. :param file_path: The path to the file relative to the upload.
- Raises
KeyError – If the file does not exist.
Restricted – If the file is restricted and upload access evaluated to False.
-
raw_file_size(file_path: str) → int¶ Returns: The size of the given raw file.
-
raw_file_object(file_path: str) → nomad.files.PathObject¶
-
write_archive(calc_id: str, data: Any) → int¶ Writes the data as archive file and returns the archive file size.
-
read_archive(calc_id: str, access: str = None) → nomad.archive.storage.ArchiveReader¶ Returns an
nomad.archive.ArchiveReaderthat contains the given calc_id. Both restricted and public archive are searched by default. The optionalaccessparameter can be used to limit this lookup to thepublicorrestrictedarchive.
-
archive_file_object(calc_id: str) → nomad.files.PathObject¶
-
add_rawfiles(path: str, move: bool = False, prefix: str = None, force_archive: bool = False, target_dir: nomad.files.DirectoryObject = None) → None¶ Add rawfiles to the upload. The given file will be copied, moved, or extracted.
- Parameters
path – Path to a directory, file, or zip file. Zip files will be extracted.
move – Whether the file should be moved instead of copied. Zips will be extracted and then deleted.
prefix – Optional path prefix for the added files.
force_archive – Expect the file to be a zip or other support archive file. Usually those files are only extracted if they can be extracted and copied instead.
target_dir – Overwrite the used directory to extract to. Default is the raw directory of this upload.
-
property
is_frozen¶ Returns True if this upload is already bagged.
-
pack(entries: Iterable[nomad.datamodel.datamodel.EntryMetadata], target_dir: nomad.files.DirectoryObject = None, skip_raw: bool = False, skip_archive: bool = False) → None¶ Replaces the staging upload data with a public upload record by packing all data into files. It is only available if upload is_bag. This is potentially a long running operation.
- Parameters
upload – The upload with all calcs and calculation metadata of the upload used to determine what files to pack and what the embargo situation is.
target_dir – optional DirectoryObject to override where to put the files. Default is the corresponding public upload files directory.
skip_raw – determine to not pack the raw data, only archive and user metadata
skip_raw – determine to not pack the archive data, only raw and user metadata
-
raw_file_manifest(path_prefix: str = None) → Generator[str, None, None]¶ Returns the path for all raw files in the archive (with a given prefix). :param path_prefix: An optional prefix; only returns those files that have the prefix.
- Returns
An iterable over all (matching) raw files.
-
raw_file_list(directory: str) → List[Tuple[str, int]]¶ Gives a list of directory contents and its size. :param directory: The directory to list
- Returns
A list of tuples with file name and size.
-
calc_files(mainfile: str, with_mainfile: bool = True, with_cutoff: bool = True) → Iterable[str]¶ Returns all the auxfiles and mainfile for a given mainfile. This implements nomad’s logic about what is part of a calculation and what not. The mainfile is first entry, the rest is sorted. :param mainfile: The mainfile relative to upload :param with_mainfile: Do include the mainfile, default is True
-
calc_id(mainfile: str) → str¶ Calculates a id for the given calc. :param mainfile: The mainfile path relative to the upload that identifies the calc in the folder structure.
- Returns
The calc id
- Raises
KeyError – If the mainfile does not exist.
-
calc_hash(mainfile: str) → str¶ Calculates a hash for the given calc based on file contents and aux file contents. :param mainfile: The mainfile path relative to the upload that identifies the calc in the folder structure.
- Returns
The calculated hash
- Raises
KeyError – If the mainfile does not exist.
-
delete() → None¶
-
-
class
nomad.files.ArchiveBasedStagingUploadFiles(upload_id: str, upload_path: str, *args, **kwargs)¶ StagingUploadFilesbased on a single uploaded archive file (.zip)- Parameters
upload_path – The path to the uploaded file.
-
__init__(upload_id: str, upload_path: str, *args, **kwargs) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
property
is_valid¶
-
extract() → None¶
-
add_rawfiles(path: str, move: bool = False, prefix: str = None, force_archive: bool = False, target_dir: nomad.files.DirectoryObject = None) → None¶ Add rawfiles to the upload. The given file will be copied, moved, or extracted.
- Parameters
path – Path to a directory, file, or zip file. Zip files will be extracted.
move – Whether the file should be moved instead of copied. Zips will be extracted and then deleted.
prefix – Optional path prefix for the added files.
force_archive – Expect the file to be a zip or other support archive file. Usually those files are only extracted if they can be extracted and copied instead.
target_dir – Overwrite the used directory to extract to. Default is the raw directory of this upload.
-
class
nomad.files.PublicUploadFilesBasedStagingUploadFiles(public_upload_files: nomad.files.PublicUploadFiles, *args, **kwargs)¶ StagingUploadFilesbased on a single uploaded archive file (.zip)- Parameters
upload_path – The path to the uploaded file.
-
__init__(public_upload_files: nomad.files.PublicUploadFiles, *args, **kwargs) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
extract(include_archive: bool = False) → None¶
-
add_rawfiles(*args, **kwargs) → None¶ Add rawfiles to the upload. The given file will be copied, moved, or extracted.
- Parameters
path – Path to a directory, file, or zip file. Zip files will be extracted.
move – Whether the file should be moved instead of copied. Zips will be extracted and then deleted.
prefix – Optional path prefix for the added files.
force_archive – Expect the file to be a zip or other support archive file. Usually those files are only extracted if they can be extracted and copied instead.
target_dir – Overwrite the used directory to extract to. Default is the raw directory of this upload.
-
pack(entries: Iterable[nomad.datamodel.datamodel.EntryMetadata], *args, **kwargs) → None¶ Packs only the archive contents and stores it in the existing public upload files.
-
class
nomad.files.PublicUploadFiles(*args, **kwargs)¶ -
__init__(*args, **kwargs) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
close()¶ Release possibly held system resources (e.g. file handles).
-
to_staging_upload_files(create: bool = False, **kwargs) → nomad.files.StagingUploadFiles¶ Casts to or creates corresponding staging upload files or returns None.
-
add_metadata_file(metadata: dict)¶
-
property
public_raw_data_file¶
-
raw_file(file_path: str, *args, **kwargs) → IO¶ Opens a raw file and returns a file-like object. Additional args, kwargs are delegated to the respective open call. :param file_path: The path to the file relative to the upload.
- Raises
KeyError – If the file does not exist.
Restricted – If the file is restricted and upload access evaluated to False.
-
raw_file_size(file_path: str) → int¶ Returns: The size of the given raw file.
-
raw_file_manifest(path_prefix: str = None) → Generator[str, None, None]¶ Returns the path for all raw files in the archive (with a given prefix). :param path_prefix: An optional prefix; only returns those files that have the prefix.
- Returns
An iterable over all (matching) raw files.
-
raw_file_list(directory: str) → List[Tuple[str, int]]¶ Gives a list of directory contents and its size. :param directory: The directory to list
- Returns
A list of tuples with file name and size.
-
read_archive(calc_id: str, access: str = None) → Any¶ Returns an
nomad.archive.ArchiveReaderthat contains the given calc_id. Both restricted and public archive are searched by default. The optionalaccessparameter can be used to limit this lookup to thepublicorrestrictedarchive.
-
re_pack(entries: Iterable[nomad.datamodel.datamodel.EntryMetadata], skip_raw: bool = False, skip_archive: bool = False) → None¶ Replaces the existing public/restricted data file pairs with new ones, based on current restricted information in the metadata. Should be used after updating the restrictions on calculations. This is potentially a long running operation.
-
nomad.archive¶
The archive storage is made from two tiers. First the whole archive is stored in files, secondly parts of the archive are stored in mongodb documents.
The file storage is done in msg-pack files. Each file contains the archive of many
entries (all entries of an upload). These msg-pack files contain the JSON serialized
version of the metainfo archive (see module:nomad.metainfo). In addition msg-pack
contains TOC information for quicker access of individual sections. See write_archive()
and read_archvive(). In addition there query functionality to partially read
specified sections from an archive: func:query_archive.
The mongo storage uses mongodb’s native bson to store JSON serialized metainfo archive data. Each document in mongodb holds the partial archive of single entry. Which parts of an archive are stored in mongo is determined by the metainfo and section annotations/categories.
nomad.doi¶
This module contains all functions necessary to manage DOI via datacite.org and its MDS API (https://support.datacite.org/docs/mds-api-guide).
-
nomad.doi.edit_url(doi: str, url: str = None)¶ Changes the URL of an already findable DOI.
-
class
nomad.doi.DOI(*args, **values)¶ -
doi¶ A unicode string field.
-
url¶ A unicode string field.
-
metadata_url¶ A unicode string field.
-
doi_url¶ A unicode string field.
-
state¶ A unicode string field.
-
create_time¶ Datetime field.
Uses the python-dateutil library if available alternatively use time.strptime to parse the dates. Note: python-dateutil’s parser is fully featured and when installed you can utilise it to convert varying types of date formats into valid python datetime objects.
Note: To default the field to the current datetime, use: DateTimeField(default=datetime.utcnow)
- Note: Microseconds are rounded to the nearest millisecond.
Pre UTC microsecond support is effectively broken. Use
ComplexDateTimeFieldif you need accurate microsecond support.
-
metadata_xml¶ A unicode string field.
-
static
create(title: str, user: nomad.datamodel.datamodel.User) → nomad.doi.DOI¶ Creates a unique DOI with the NOMAD DOI prefix.
-
create_draft()¶
-
delete(*args, **kwargs)¶ Delete the
Documentfrom the database. This will only take effect if the document has been previously saved.- Parameters
signal_kwargs – (optional) kwargs dictionary to be passed to the signal calls.
write_concern – Extra keyword arguments are passed down which will be used as options for the resultant
getLastErrorcommand. For example,save(..., w: 2, fsync: True)will wait until at least two servers have recorded the write and will force an fsync on the primary server.
Changed in version 0.10.7: Add signal_kwargs argument
-
make_findable()¶
-
exception
DoesNotExist¶
-
exception
MultipleObjectsReturned¶
-
id¶ A unicode string field.
-
objects¶ The default QuerySet Manager.
Custom QuerySet Manager functions can extend this class and users can add extra queryset functionality. Any custom manager methods must accept a
Documentclass as its first argument, and aQuerySetas its second argument.The method function should return a
QuerySet, probably the same one that was passed in, but modified in some way.
-
nomad.parsing¶
The parsing module is an interface for the existing NOMAD-coe parsers. This module redefines some of the old NOMAD-coe python-common functionality to create a more coherent interface to the parsers.
Assumption about parsers¶
For now, we make a few assumption about parsers - they always work on the same meta-info version - they have no conflicting python requirements - they can be loaded at the same time and can be used within the same python process - they are uniquely identified by a GIT URL and publicly accessible - their version is uniquely identified by a GIT commit SHA
Each parser is defined via an instance of Parser. The implementation LegacyParser is used for most NOMAD-coe parsers.
-
class
nomad.parsing.Parser¶ Instances specify a parser. It allows to find main files from given uploaded and extracted files. Further, allows to run the parser on those ‘main files’.
-
name= 'parsers/parser'¶
-
__init__()¶ Initialize self. See help(type(self)) for accurate signature.
-
abstract
is_mainfile(filename: str, mime: str, buffer: bytes, decoded_buffer: str, compression: str = None) → bool¶ Checks if a file is a mainfile for the parsers.
- Parameters
filename – The filesystem path to the mainfile
mime – The mimetype of the mainfile guessed with libmagic
buffer – The first 2k of the mainfile contents
compression – The compression of the mainfile
[None, 'gz', 'bz2']
-
abstract
parse(mainfile: str, archive: nomad.datamodel.datamodel.EntryArchive, logger=None) → None¶ Runs the parser on the given mainfile and populates the result in the given archive root_section. It allows to be run repeatedly for different mainfiles.
- Parameters
mainfile – A path to a mainfile that this parser can parse.
archive – An instance of the section
EntryArchive. It might contain asection_metadatawith information about the entry.logger – A optional logger
-
classmethod
main(mainfile)¶
-
The are sub-classes for parsers with special purposes.
-
class
nomad.parsing.Parser Instances specify a parser. It allows to find main files from given uploaded and extracted files. Further, allows to run the parser on those ‘main files’.
-
__init__() Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.MatchingParser(name: str, code_name: str, code_homepage: str = None, mainfile_contents_re: str = None, mainfile_binary_header: bytes = None, mainfile_binary_header_re: bytes = None, mainfile_mime_re: str = 'text/.*', mainfile_name_re: str = '.*', mainfile_alternative: bool = False, domain='dft', supported_compressions: List[str] = [])¶ A parser implementation that uses regular expressions to match mainfiles.
- Parameters
code_name – The name of the code or input format
code_homepage – The homepage of the code or input format
mainfile_mime_re – A regexp that is used to match against a files mime type
mainfile_contents_re – A regexp that is used to match the first 1024 bytes of a potential mainfile.
mainfile_name_re – A regexp that is used to match the paths of potential mainfiles
mainfile_alternative – If True files are mainfile if no mainfile_name_re matching file is present in the same directory.
domain – The domain that this parser should be used for. Default is ‘dft’.
supported_compressions – A list of [gz, bz2], if the parser supports compressed files
-
__init__(name: str, code_name: str, code_homepage: str = None, mainfile_contents_re: str = None, mainfile_binary_header: bytes = None, mainfile_binary_header_re: bytes = None, mainfile_mime_re: str = 'text/.*', mainfile_name_re: str = '.*', mainfile_alternative: bool = False, domain='dft', supported_compressions: List[str] = []) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
class
nomad.parsing.MissingParser(*args, **kwargs)¶ A parser implementation that just fails and is used to match mainfiles with known patterns of corruption.
-
__init__(*args, **kwargs)¶ Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.BrokenParser(*args, **kwargs)¶ A parser implementation that just fails and is used to match mainfiles with known patterns of corruption.
-
__init__(*args, **kwargs)¶ Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.TemplateParser(*args, **kwargs)¶ A parser that generates data based on a template given via the mainfile. The template is basically some archive json. Only
-
__init__(*args, **kwargs)¶ Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.GenerateRandomParser(*args, **kwargs)¶ -
__init__(*args, **kwargs)¶ Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.ChaosParser¶ Parser that emulates typical error situations. Files can contain a json string (or object with key chaos) with one of the following string values: - exit - deadlock - consume_ram - exception - segfault - random
-
__init__()¶ Initialize self. See help(type(self)) for accurate signature.
-
-
class
nomad.parsing.EmptyParser(name: str, code_name: str, code_homepage: str = None, mainfile_contents_re: str = None, mainfile_binary_header: bytes = None, mainfile_binary_header_re: bytes = None, mainfile_mime_re: str = 'text/.*', mainfile_name_re: str = '.*', mainfile_alternative: bool = False, domain='dft', supported_compressions: List[str] = [])¶ Implementation that produces an empty code_run
-
__init__(name: str, code_name: str, code_homepage: str = None, mainfile_contents_re: str = None, mainfile_binary_header: bytes = None, mainfile_binary_header_re: bytes = None, mainfile_mime_re: str = 'text/.*', mainfile_name_re: str = '.*', mainfile_alternative: bool = False, domain='dft', supported_compressions: List[str] = []) → None¶ Initialize self. See help(type(self)) for accurate signature.
-
The implementation LegacyParser is used for most NOMAD-coe parsers.
The parser definitions are available via the following two variables.
-
nomad.parsing.parsers.parsers= [<nomad.parsing.artificial.GenerateRandomParser object>, <nomad.parsing.artificial.TemplateParser object>, <nomad.parsing.artificial.ChaosParser object>, parsers/phonopy, parsers/vasp, parsers/exciting, parsers/fhi-aims, parsers/fhi-vibes, parsers/cp2k, parsers/crystal, parsers/cpmd, parsers/nwchem, parsers/bigdft, parsers/wien2k, parsers/band, parsers/quantumespresso, parsers/gaussian, parsers/abinit, parsers/orca, parsers/castep, parsers/dl-poly, parsers/lib-atoms, parsers/octopus, parsers/gpaw, parsers/atk, parsers/gulp, parsers/siesta, parsers/elk, parsers/elastic, parsers/gamess, parsers/turbomole, parsers/mpes, parsers/aptfim, parsers/eels, parsers/qbox, parsers/dmol, parsers/fleur, parsers/molcas, parsers/onetep, parsers/openkim, parsers/tinker, parsers/lammps, parsers/amber, parsers/gromacs, parsers/lobster, parsers/gromos, parsers/namd, parsers/charmm, parsers/dftbplus, parsers/asap, parsers/fplo, parsers/mopac, parsers/openmx, parsers/archive, <nomad.parsing.parser.BrokenParser object>]¶ Built-in mutable sequence.
If no argument is given, the constructor creates a new empty list. The argument must be an iterable if specified.
-
nomad.parsing.parsers.parser_dict= {'missing/crystal': missing/crystal, 'missing/fhi-aims': missing/fhi-aims, 'missing/octopus': missing/octopus, 'missing/wien2k': missing/wien2k, 'parser/broken': <nomad.parsing.parser.BrokenParser object>, 'parser/fleur': parsers/fleur, 'parser/molcas': parsers/molcas, 'parser/octopus': parsers/octopus, 'parser/onetep': parsers/onetep, 'parsers/abinit': parsers/abinit, 'parsers/amber': parsers/amber, 'parsers/aptfim': parsers/aptfim, 'parsers/archive': parsers/archive, 'parsers/asap': parsers/asap, 'parsers/atk': parsers/atk, 'parsers/band': parsers/band, 'parsers/bigdft': parsers/bigdft, 'parsers/broken': <nomad.parsing.parser.BrokenParser object>, 'parsers/castep': parsers/castep, 'parsers/chaos': <nomad.parsing.artificial.ChaosParser object>, 'parsers/charmm': parsers/charmm, 'parsers/cp2k': parsers/cp2k, 'parsers/cpmd': parsers/cpmd, 'parsers/crystal': parsers/crystal, 'parsers/dftbplus': parsers/dftbplus, 'parsers/dl-poly': parsers/dl-poly, 'parsers/dmol': parsers/dmol, 'parsers/eels': parsers/eels, 'parsers/elastic': parsers/elastic, 'parsers/elk': parsers/elk, 'parsers/exciting': parsers/exciting, 'parsers/fhi-aims': parsers/fhi-aims, 'parsers/fhi-vibes': parsers/fhi-vibes, 'parsers/fleur': parsers/fleur, 'parsers/fplo': parsers/fplo, 'parsers/gamess': parsers/gamess, 'parsers/gaussian': parsers/gaussian, 'parsers/gpaw': parsers/gpaw, 'parsers/gromacs': parsers/gromacs, 'parsers/gromos': parsers/gromos, 'parsers/gulp': parsers/gulp, 'parsers/lammps': parsers/lammps, 'parsers/lib-atoms': parsers/lib-atoms, 'parsers/lobster': parsers/lobster, 'parsers/molcas': parsers/molcas, 'parsers/mopac': parsers/mopac, 'parsers/mpes': parsers/mpes, 'parsers/namd': parsers/namd, 'parsers/nwchem': parsers/nwchem, 'parsers/octopus': parsers/octopus, 'parsers/onetep': parsers/onetep, 'parsers/openkim': parsers/openkim, 'parsers/openmx': parsers/openmx, 'parsers/orca': parsers/orca, 'parsers/phonopy': parsers/phonopy, 'parsers/qbox': parsers/qbox, 'parsers/quantumespresso': parsers/quantumespresso, 'parsers/random': <nomad.parsing.artificial.GenerateRandomParser object>, 'parsers/siesta': parsers/siesta, 'parsers/template': <nomad.parsing.artificial.TemplateParser object>, 'parsers/tinker': parsers/tinker, 'parsers/turbomole': parsers/turbomole, 'parsers/vasp': parsers/vasp, 'parsers/wien2k': parsers/wien2k}¶ A dict to access parsers by name. Usually ‘parsers/<…>’, e.g. ‘parsers/vasp’.
Parsers are reused for multiple calculations.
Parsers and calculation files are matched via regular expressions.
-
nomad.parsing.parsers.match_parser(mainfile_path: str, strict=True) → nomad.parsing.parser.Parser¶ Performs parser matching. This means it take the given mainfile and potentially opens it with the given callback and tries to identify a parser that can parse the file.
This is determined by filename (e.g. .out), mime type (e.g. text/, application/xml), and beginning file contents.
- Parameters
mainfile_path – Path to the mainfile
strict – Only match strict parsers, e.g. no artificial parsers for missing or empty entries.
Returns: The parser, or None if no parser could be matched.
Parsers in NOMAD-coe use a backend to create output. There are different NOMAD-coe basends. In nomad@FAIRDI, we only currently only use a single backed. The following classes provide a interface definition for backends as an ABC and a concrete implementation based on nomad@fairdi’s metainfo:
nomad.normalizing¶
After parsing calculations have to be normalized with a set of normalizers. In NOMAD-coe those were programmed in python (we’ll reuse) and scala (we’ll rewrite).
Currently the normalizers are: - system.py (contains aspects of format stats, system, system type, and symmetry normalizer) - optimade.py - fhiaims.py - dos.py - encyclopedia.py (used to create the data in NOMAD-coe Encyclopedia)
The normalizers are available via
-
nomad.normalizing.normalizers: Iterable[Type[nomad.normalizing.normalizer.Normalizer]] = [<class 'nomad.normalizing.system.SystemNormalizer'>, <class 'nomad.normalizing.optimade.OptimadeNormalizer'>, <class 'nomad.normalizing.dos.DosNormalizer'>, <class 'nomad.normalizing.band_structure.BandStructureNormalizer'>, <class 'nomad.normalizing.workflow.WorkflowNormalizer'>, <class 'nomad.normalizing.encyclopedia.encyclopedia.EncyclopediaNormalizer'>]¶ Built-in mutable sequence.
If no argument is given, the constructor creates a new empty list. The argument must be an iterable if specified.
There is one ABC for all normalizer:
nomad.processing¶
Processing comprises everything that is necessary to take an uploaded user file, processes it, and store all necessary data for repository, archive, and potential future services (e.g. encyclopedia).
Processing is build on top of celery (http://www.celeryproject.org/) and
mongodb (http://www.mongodb.org).
Celery provides a task-based programming model for distributed computing. It uses
a broker, e.g. a distributed task queue like RabbitMQ to distribute tasks. We
use mongodb to store the current state of processing in Upload and
Calculation documents. This combination allows us to easily distribute
processing work while having the processing state, i.e. (intermediate) results,
always available.
This module is structured into our celery app and abstract process base class
Proc (base.py), and the concrete processing classes
Upload and Calc (data.py).
This module does not contain the functions to do the actual work. Those are encapsulated
in nomad.files, nomad.repo, nomad.users,
nomad.parsing, and nomad.normalizing.
Refer to http://www.celeryproject.org/ to learn about celery apps and workers. The nomad celery app uses a RabbitMQ broker. We use celery to distribute processing load in a cluster.
We use an abstract processing base class and document (Proc) that provides all
necessary functions to execute a process as a series of potentially distributed steps. In
addition the processing state is persisted in mongodb using mongoengine. Instead of
exchanging serialized state between celery tasks, we use the mongodb documents to
exchange data. Therefore, the mongodb always contains the latest processing state.
We also don’t have to deal with celery result backends and synchronizing with them.
-
class
nomad.processing.base.Proc(*args, **values)¶ Base class for objects that are involved in processing and need persistent processing state.
It solves two issues. First, distributed operation (via celery) and second keeping state of a chain of potentially failing processing tasks. Both are controlled via decorators @process and @task. Subclasses should use these decorators on their methods. Parameters are not supported for decorated functions. Use fields on the document instead.
Processing state will be persistet at appropriate times and must not be persistet manually. All attributes are stored to mongodb.
Possible processing states are PENDING, RUNNING, FAILURE, and SUCCESS.
-
current_task¶ the currently running or last completed task
-
tasks_status¶ the overall status of the processing
-
errors¶ a list of errors that happened during processing. Error fail a processing run
-
warnings¶ a list of warnings that happened during processing. Warnings do not fail a processing run
-
create_time¶ the time of creation (not the start of processing)
-
complete_time¶ the time that processing completed (successfully or not)
-
current_process¶ the currently or last run asyncronous process
-
process_status¶ the status of the currently or last run asyncronous process
-
__init__(*args, **values)¶ Initialise a document or an embedded document.
- Parameters
values – A dictionary of keys and values for the document. It may contain additional reserved keywords, e.g. “__auto_convert”.
__auto_convert – If True, supplied values will be converted to Python-type values via each field’s to_python method.
__only_fields – A set of fields that have been loaded for this document. Empty if all fields have been loaded.
_created – Indicates whether this is a brand new document or whether it’s already been persisted before. Defaults to true.
-
There are two concrete processes Upload and :class: Calc. Instances of both
classes do represent the processing state, as well as the respective entity.
-
class
nomad.processing.data.Upload(**kwargs)¶ Represents uploads in the databases. Provides persistence access to the files storage, and processing state.
-
name¶ Optional user provided upload name.
-
upload_path¶ The fs path were the uploaded files was stored during upload.
-
temporary¶ True if the uploaded file should be removed after extraction.
-
upload_id¶ The upload id generated by the database or the uploaded NOMAD deployment.
-
upload_time¶ Datetime of the original upload independent of the NOMAD deployment it was first uploaded to.
-
user_id¶ The id of the user that created this upload.
-
published¶ Boolean that indicates that the upload is published on this NOMAD deployment.
-
publish_time¶ Datetime when the upload was initially published on this NOMAD deployment.
-
last_update¶ Datetime of the last modifying process run (publish, re-processing, upload).
-
publish_directly¶ Boolean indicating that this upload should be published after initial processing.
-
from_oasis¶ Boolean indicating that this upload is coming from another NOMAD deployment.
-
oasis_id¶ The deployment id of the NOMAD that uploaded the upload.
-
published_to¶ A list of deployment ids where this upload has been successfully uploaded to.
-
joined¶ Boolean indicates if the running processing has joined (
check_join()).
-
id_field= 'upload_id'¶
-
upload_id A unicode string field.
-
upload_path A unicode string field.
-
temporary Boolean field type.
New in version 0.1.2.
-
embargo_length¶ 32-bit integer field.
-
name A unicode string field.
-
upload_time Datetime field.
Uses the python-dateutil library if available alternatively use time.strptime to parse the dates. Note: python-dateutil’s parser is fully featured and when installed you can utilise it to convert varying types of date formats into valid python datetime objects.
Note: To default the field to the current datetime, use: DateTimeField(default=datetime.utcnow)
- Note: Microseconds are rounded to the nearest millisecond.
Pre UTC microsecond support is effectively broken. Use
ComplexDateTimeFieldif you need accurate microsecond support.
-
user_id A unicode string field.
-
published Boolean field type.
New in version 0.1.2.
-
publish_time Datetime field.
Uses the python-dateutil library if available alternatively use time.strptime to parse the dates. Note: python-dateutil’s parser is fully featured and when installed you can utilise it to convert varying types of date formats into valid python datetime objects.
Note: To default the field to the current datetime, use: DateTimeField(default=datetime.utcnow)
- Note: Microseconds are rounded to the nearest millisecond.
Pre UTC microsecond support is effectively broken. Use
ComplexDateTimeFieldif you need accurate microsecond support.
-
last_update Datetime field.
Uses the python-dateutil library if available alternatively use time.strptime to parse the dates. Note: python-dateutil’s parser is fully featured and when installed you can utilise it to convert varying types of date formats into valid python datetime objects.
Note: To default the field to the current datetime, use: DateTimeField(default=datetime.utcnow)
- Note: Microseconds are rounded to the nearest millisecond.
Pre UTC microsecond support is effectively broken. Use
ComplexDateTimeFieldif you need accurate microsecond support.
-
from_oasis Boolean field type.
New in version 0.1.2.
-
oasis_deployment_id¶ A unicode string field.
-
published_to A list field that wraps a standard field, allowing multiple instances of the field to be used as a list in the database.
If using with ReferenceFields see: one-to-many-with-listfields
Note
Required means it cannot be empty - as the default for ListFields is []
-
joined Boolean field type.
New in version 0.1.2.
-
meta: Any = None¶
-
__init__(**kwargs)¶ Initialise a document or an embedded document.
- Parameters
values – A dictionary of keys and values for the document. It may contain additional reserved keywords, e.g. “__auto_convert”.
__auto_convert – If True, supplied values will be converted to Python-type values via each field’s to_python method.
__only_fields – A set of fields that have been loaded for this document. Empty if all fields have been loaded.
_created – Indicates whether this is a brand new document or whether it’s already been persisted before. Defaults to true.
-
publish_directly Boolean field type.
New in version 0.1.2.
-
metadata_file_cached¶
-
property
metadata¶ Getter, setter for user metadata. Metadata is pickled to and from the public bucket to allow sharing among all processes. Usually uploads do not have (much) user defined metadata, but users provide all metadata per upload as part of the publish process. This will change, when we introduce editing functionality and metadata will be provided through different means.
-
classmethod
get(id: str, include_published: bool = True) → nomad.processing.data.Upload¶
-
classmethod
user_uploads(user: nomad.datamodel.datamodel.User, **kwargs) → List[nomad.processing.data.Upload]¶ Returns all uploads for the given user. Kwargs are passed to mongo query.
-
property
uploader¶
-
get_logger(**kwargs)¶
-
classmethod
create(**kwargs) → nomad.processing.data.Upload¶ Creates a new upload for the given user, a user given name is optional. It will populate the record with a signed url and pending
UploadProc. The upload will be already saved to the database.- Parameters
user – The user that created the upload.
-
delete()¶ Deletes this upload process state entry and its calcs.
-
delete_upload_local()¶ Deletes the upload, including its processing state and staging files. Local version without celery processing.
-
delete_upload()¶ Deletes the upload, including its processing state and staging files. This starts the celery process of deleting the upload.
-
publish_upload()¶ Moves the upload out of staging to the public area. It will pack the staging upload files in to public upload files.
-
publish_from_oasis()¶ Uploads the already published upload to a different NOMAD deployment. This allows to push uploads from an OASIS to the central NOMAD.
-
re_process_upload()¶ A process that performs the re-processing of a earlier processed upload.
Runs the distributed process of fully reparsing/re-normalizing an existing and already published upload. Will renew the archive part of the upload and update mongo and elastic search entries.
TODO this implementation does not do any re-matching. This will be more complex due to handling of new or missing matches.
-
re_pack()¶ A process that repacks the raw and archive data based on the current embargo data.
-
process_upload()¶ A process that performs the initial upload processing.
-
uploading()¶ A no-op task as a stand-in for receiving upload data.
-
property
upload_files¶
-
property
staging_upload_files¶
-
extracting()¶ The task performed before the actual parsing/normalizing: extracting the uploaded files.
-
match_mainfiles() → Iterator[Tuple[str, object]]¶ Generator function that matches all files in the upload to all parsers to determine the upload’s mainfiles.
- Returns
Tuples of mainfile, filename, and parsers
-
parse_all()¶ The task used to identify mainfile/parser combinations among the upload’s files, creates respective
Calcinstances, and triggers their processing.
-
on_process_complete(process_name)¶ Callback that is called when the corrent process completed
-
check_join()¶ Performs an evaluation of the join condition and triggers the
cleanup()task if necessary. The join condition allows to run thecleanupafter all calculations have been processed. The upload processing stops after all calculation processing have been triggered (parse_all()orre_process_upload()). The cleanup task is then run within the last calculation process (the one that triggered the join by calling this method).
-
reset(force=False)¶ Resets the task chain. Assumes there no current running process.
-
classmethod
reset_pymongo_update(worker_hostname: str = None)¶ Returns a pymongo update dict part to reset calculations.
-
cleanup()¶ The task that “cleans” the processing, i.e. removed obsolete files and performs pending archival operations. Depends on the type of processing.
-
get_calc(calc_id) → nomad.processing.data.Calc¶ Returns the upload calc with the given id or
None.
-
property
processed_calcs¶ The number of successfully or not successfully processed calculations. I.e. calculations that have finished processing.
-
property
total_calcs¶ The number of all calculations.
-
property
failed_calcs¶ The number of calculations with failed processing.
-
property
pending_calcs¶ The number of calculations with pending processing.
-
all_calcs(start, end, order_by=None)¶ Returns all calculations, paginated and ordered.
- Parameters
start – the start index of the requested page
end – the end index of the requested page
order_by – the property to order by
-
property
outdated_calcs¶ All successfully processed and outdated calculations.
-
property
calcs¶ All successfully processed calculations.
-
entries_metadata(user_metadata: dict = None) → Iterator[Iterable[nomad.datamodel.datamodel.EntryMetadata]]¶ This is the
nomad.datamodeltransformation method to transform processing upload’s entries into list ofnomad.datamodel.EntryMetadataobjects.- Parameters
user_metadata – A dict of user metadata that is applied to the resulting datamodel data and the respective calculations.
-
entry_ids() → Iterable[str]¶
-
exception
DoesNotExist¶
-
exception
MultipleObjectsReturned¶
-
id¶ A unicode string field.
-
objects¶ The default QuerySet Manager.
Custom QuerySet Manager functions can extend this class and users can add extra queryset functionality. Any custom manager methods must accept a
Documentclass as its first argument, and aQuerySetas its second argument.The method function should return a
QuerySet, probably the same one that was passed in, but modified in some way.
-
tasks= ['uploading', 'extracting', 'parse_all', 'cleanup']¶
-
user_metadata() → Iterable[nomad.datamodel.datamodel.EntryMetadata]¶
-
compress_and_set_metadata(metadata: Dict[str, Any]) → None¶ Stores the given user metadata in the upload document. This is the metadata adhering to the API model (
UploadMetaData). Most quantities can be stored for the upload and for each calculation. This method will try to move same values from the calculation to the upload to “compress” the data.
-
-
class
nomad.processing.data.Calc(*args, **kwargs)¶ Instances of this class represent calculations. This class manages the elastic search index entry, files, and archive for the respective calculation.
It also contains the calculations processing and its state.
The attribute list, does not include the various metadata properties generated while parsing, including
code_name,code_version, etc.-
calc_id¶ the calc_id of this calc
-
parser¶ the name of the parser used to process this calc
-
upload_id¶ the id of the upload used to create this calculation
-
mainfile¶ the mainfile (including path in upload) that was used to create this calc
-
metadata¶ the metadata record wit calc and user metadata, see
datamodel.EntryMetadata
-
calc_id A unicode string field.
-
upload_id A unicode string field.
-
mainfile A unicode string field.
-
parser A unicode string field.
-
metadata A dictionary field that wraps a standard Python dictionary. This is similar to an embedded document, but the structure is not defined.
Note
Required means it cannot be empty - as the default for DictFields is {}
New in version 0.3.
Changed in version 0.5: - Can now handle complex / varying types of data
-
meta: Any = None¶
-
__init__(*args, **kwargs)¶ Initialise a document or an embedded document.
- Parameters
values – A dictionary of keys and values for the document. It may contain additional reserved keywords, e.g. “__auto_convert”.
__auto_convert – If True, supplied values will be converted to Python-type values via each field’s to_python method.
__only_fields – A set of fields that have been loaded for this document. Empty if all fields have been loaded.
_created – Indicates whether this is a brand new document or whether it’s already been persisted before. Defaults to true.
-
classmethod
get(id)¶
-
property
entry_id¶ Just an alias for calc_id.
-
property
mainfile_file¶
-
property
upload¶
-
apply_entry_metadata(entry_metadata: nomad.datamodel.datamodel.EntryMetadata)¶
-
create_metadata() → nomad.datamodel.datamodel.EntryMetadata¶ Returns a
nomad.datamodel.EntryMetadatawith values from this processing object, not necessarily the user metadata nor the metadata from the archive.
-
entry_metadata(upload_files: nomad.files.UploadFiles) → nomad.datamodel.datamodel.EntryMetadata¶ Returns a complete set of
nomad.datamodel.EntryMetadataincluding the user metadata and metadata from the archive.- Parameters
upload_files – The
nomad.files.UploadFilesinstance to read the archive from.cache – A boolean that indicates if the archive file should be left unclosed, e.g. if this method is called for many entries of the same upload.
-
user_metadata() → nomad.datamodel.datamodel.EntryMetadata¶ Returns a
nomad.datamodel.EntryMetadatawith values from this processing object and the user metadata, not necessarily the metadata from the archive.
-
property
upload_files¶
-
get_logger(**kwargs)¶ Returns a wrapped logger that additionally saves all entries to the calculation processing log in the archive.
-
re_process_calc()¶ Processes a calculation again. This means there is already metadata and instead of creating it initially, we are just updating the existing records.
-
process_calc()¶ Processes a new calculation that has no prior records in the mongo, elastic, or filesystem storage. It will create an initial set of (user) metadata.
-
on_fail()¶
-
on_process_complete(process_name)¶ Callback that is called when the corrent process completed
-
parsing()¶ The task that encapsulates all parsing related actions.
-
process_phonon()¶ Function that is run for phonon calculation before cleanup. This task is run by the celery process that is calling the join for the upload.
This function re-opens the Archive for this calculation to add method information from another referenced archive. Updates the method information in section_encyclopedia as well as the DFT domain metadata.
-
normalizing()¶ The task that encapsulates all normalizing related actions.
-
archiving()¶ The task that encapsulates all archival related actions.
-
write_archive(archive: nomad.datamodel.datamodel.EntryArchive)¶
-
exception
DoesNotExist¶
-
exception
MultipleObjectsReturned¶
-
id¶ A unicode string field.
-
objects¶ The default QuerySet Manager.
Custom QuerySet Manager functions can extend this class and users can add extra queryset functionality. Any custom manager methods must accept a
Documentclass as its first argument, and aQuerySetas its second argument.The method function should return a
QuerySet, probably the same one that was passed in, but modified in some way.
-
tasks= ['parsing', 'normalizing', 'archiving']¶
-
nomad.search¶
This module represents calculations in elastic search.
-
exception
nomad.search.AlreadyExists¶
-
exception
nomad.search.ElasticSearchError¶
-
exception
nomad.search.AuthenticationRequiredError¶
-
exception
nomad.search.ScrollIdNotFound¶
-
exception
nomad.search.InvalidQuery¶
-
nomad.search.delete_upload(upload_id)¶ Delete all entries with given
upload_idfrom the index.
-
nomad.search.delete_entry(calc_id)¶ Delete the entry with the given
calc_idfrom the index.
-
nomad.search.publish(calcs: Iterable[nomad.datamodel.datamodel.EntryMetadata]) → None¶ Update all given calcs with their metadata and set
publish = True.
-
nomad.search.index_all(calcs: Iterable[nomad.datamodel.datamodel.EntryMetadata], do_refresh=True) → int¶ Adds all given calcs with their metadata to the index.
- Returns
Number of failed entries.
-
nomad.search.refresh()¶
-
class
nomad.search.SearchRequest(domain: str = 'dft', query=None)¶ Represents a search request and allows to execute that request. It allows to compose the following features: a query; statistics (metrics and aggregations); quantity values; scrolling, pagination for entries; scrolling for quantity values.
The query part filters NOMAD data before the other features come into effect. There are specialized methods for configuring the
owner()andtime_range()queries. Quantity’s can be search for by setting them as attributes.The aggregations for statistics can be requested for pre-configured quantities. These bucket aggregations come with a metric calculated for each each possible quantity value.
The other possible form of aggregations, allows to get quantity values as results (e.g. get all datasets, get all users, etc.). Each value can be accompanied by metrics (over all entries with that value) and an example value.
Of course, searches can return a set of search results. Search objects can be configured with pagination or scrolling for these results. Pagination is the default and also allows ordering of results. Scrolling can be used if all entries need to be ‘scrolled through’. This might be necessary, since elastic search has limits on possible pages (e.g. ‘from’ must by smaller than 10000). On the downside, there is no ordering on scrolling.
There is also scrolling for quantities to go through all quantity values. There is no paging for aggregations.
-
__init__(domain: str = 'dft', query=None)¶ Initialize self. See help(type(self)) for accurate signature.
-
domain(domain: str = None)¶ Applies the domain of this request to the query. Allows to optionally update the domain of this request.
-
owner(owner_type: str = 'all', user_id: str = None)¶ Uses the query part of the search to restrict the results based on the owner. The possible types are:
allfor all calculations;publicfor calculations visible by everyone, excluding embargo-ed entries and entries only visible to the given user;visibleall data that is visible by the user, excluding embargo-ed entries from other users;userfor all calculations of to the given user;stagingfor all calculations in staging of the given user.- Parameters
owner_type – The type of the owner query, see above.
user_id – The ‘owner’ given as the user’s unique id.
- Raises
KeyError – If the given owner_type is not supported
ValueError – If the owner_type requires a user but none is given, or the given user is not allowed to use the given owner_type.
-
search_parameters(**kwargs)¶ Configures the existing query with additional search parameters. Kwargs are interpreted as key value pairs. Keys have to coresspond to valid entry quantities in the domain’s (DFT calculations) datamodel. Alternatively search parameters can be set via attributes.
-
search_parameter(name, value)¶
-
query(query)¶ Adds the given query as a ‘and’ (i.e. ‘must’) clause to the request.
-
query_expression(expression) → nomad.search.SearchRequest¶
-
time_range(start: datetime.datetime, end: datetime.datetime)¶ Adds a time range to the query.
-
property
q¶ The underlying elasticsearch_dsl query object
-
totals(metrics_to_use: List[str] = [])¶ Configure the request to return overall totals for the given metrics.
The statics are returned with the other quantity statistics under the pseudo quantity name ‘total’. ‘total’ contains the pseudo value ‘all’. It is used to store the metrics aggregated over all entries in the search results.
-
statistics(statistics: List[str], metrics_to_use: List[str] = [])¶ Configures the domain’s default statistics.
-
statistic(quantity_name: str, size: int, metrics_to_use: List[str] = [], order: Dict[str, str] = {'_key': 'asc'}, include: str = None)¶ This can be used to display statistics over the searched entries and allows to implement faceted search on the top values for each quantity.
The metrics contain overall and per quantity value sums of code runs (calcs), unique code runs, datasets, and additional domain specific metrics (e.g. total energies, and unique geometries for DFTcalculations). The quantities that can be aggregated to metrics are defined in module:datamodel. Aggregations and respective metrics are calculated for aggregations given in
aggregationsand metrics inaggregation_metrics. As a pseudo aggregationtotal_metricsare calculation over all search results. Theaggregationsgives tuples of quantities and default aggregation sizes.The search results will contain a dictionary
statistics. This has a key for each configured quantity. Each quantity key will hold a dict with a key for each quantity value. Each quantity value key will hold a dict with a key for each metric. The values will be the actual aggregated metric values.- Parameters
quantity_name – The quantity to aggregate statistics for. Only works on keyword field.
metrics_to_use – The metrics calculated over the aggregations. Can be
unique_code_runs,datasets, other domain specific metrics. The basic doc_count metriccode_runsis always given.order – The order dictionary is passed to the elastic search aggregation.
include – Uses an regular expression in ES to only return values that include the given substring.
-
date_histogram(metrics_to_use: List[str] = [], interval: str = '1M')¶ Adds a date histogram on the given metrics to the statistics part.
-
quantities(**kwargs)¶ Shorthand for adding multiple quantities. See
quantity(). Keywork argument keys are quantity name, values are tuples of size and after value.
-
quantity(name, size=100, after=None, examples=0, examples_source=None, order_by: str = None, order: str = 'desc')¶ Adds a requests for values of the given quantity. It allows to scroll through all values via elasticsearch’s composite aggregations. The response will contain the quantity values and an example entry for each value.
This can be used to implement continues scrolling through authors, datasets, or uploads within the searched entries.
If one or more quantities are specified, the search results will contain a dictionary
quantities. The keys are quantity name the values dictionary with ‘after’ and ‘values’ key. The ‘values’ key holds a dict with all the values as keys and their entry count as values (i.e. number of entries with that value).- Parameters
name – The quantity name. Must be in
quantities.after – The ‘after’ value allows to scroll over various requests, by providing the ‘after’ value of the last search. The ‘after’ value is part of the response. Use
Nonein the first request.size – The size gives the ammount of maximum values in the next scroll window. If the size is None, a maximum of 100 quantity values will be requested.
examples – Number of results to return that has each value
order_by – A sortable quantity that should be used to order. By default, the max of each value bucket is used.
order – “desc” or “asc”
-
global_statistics()¶ Adds general statistics to the request. The results will have a key called global_statistics.
-
exclude(*args)¶ Exclude certain elastic fields from the search results.
-
include(*args)¶ Include only the given fields in the search results.
-
execute()¶ Executes without returning actual results. Only makes sense if the request was configured for statistics or quantity values.
-
execute_scan(order_by: str = None, order: int = -1, **kwargs)¶ This execute the search as scan. The result will be a generator over the found entries. Everything but the query part of this object, will be ignored.
-
execute_paginated(page: int = 1, per_page=10, page_offset: int = None, order_by: str = None, order: int = -1)¶ Executes the search and returns paginated results. Those are sorted.
- Parameters
page – The requested page, starts with 1.
per_page – The number of entries per page.
page_offset – Instead of a page number, use this absolute offset.
order_by – The quantity to order by.
order – -1 or 1 for descending or ascending order.
-
execute_scrolled(scroll_id: str = None, size: int = 1000, scroll: str = '5m', order_by: str = None, order: int = -1)¶ Executes a scrolling search. based on ES scroll API. Pagination is replaced with scrolling, no ordering is available, no statistics, no quantities will be provided.
Scrolling is done by calling this function again and again with the same
scroll_id. Each time, this function will return the next batch of search results. If thescroll_idis not available anymore, a newscroll_idis assigned and scrolling starts from the beginning again.The response will contain a ‘scroll’ part with attributes ‘total’, ‘scroll_id’, and ‘size’.
- Parameters
scroll_id – The scroll id to receive the next batch from. None will create a new scroll.
size – The batch size in number of hits.
scroll – The time the scroll should be kept alive (i.e. the time between requests to this method) in ES time units. Default is 5 minutes.
TODO support order and order_by
-
execute_aggregated(after: str = None, per_page: int = 1000, includes: List[str] = None)¶ Uses a composite aggregation on top of the search to go through the result set. This allows to go arbirarely deep without using scroll. But, it will only return results with
upload_id,calc_idand the given quantities. The results will be ‘ordered’ byupload_id.- Parameters
after – The key that determines the start of the current page. This after key is returned with each response. Use None (default) for the first request.
per_page – The size of each page.
includes – A list of quantity names that should be returned in addition to
upload_idandcalc_id.
-
-
nomad.search.flat(obj, prefix=None)¶ Helper that translates nested result objects into flattened dicts with
domain.quantityas keys.
-
nomad.search.search(owner: str = 'public', query: Union[Mapping[str, Union[str, int, float, bool, datetime.datetime, List[Union[str, int, float, bool, datetime.datetime]], nomad.app.v1.models.Lte, nomad.app.v1.models.Lt, nomad.app.v1.models.Gte, nomad.app.v1.models.Gt, nomad.app.v1.models.Any_, nomad.app.v1.models.All, nomad.app.v1.models.None_]], nomad.app.v1.models.And, nomad.app.v1.models.Or, nomad.app.v1.models.Not] = None, pagination: nomad.app.v1.models.EntryPagination = None, required: nomad.app.v1.models.MetadataRequired = None, aggregations: Dict[str, nomad.app.v1.models.Aggregation] = {}, statistics: Dict[str, nomad.app.v1.models.Statistic] = {}, user_id: str = None) → nomad.app.v1.models.SearchResponse¶
-
nomad.search.update_by_query(update_script: str, owner: str = 'public', query: Union[Mapping[str, Union[str, int, float, bool, datetime.datetime, List[Union[str, int, float, bool, datetime.datetime]], nomad.app.v1.models.Lte, nomad.app.v1.models.Lt, nomad.app.v1.models.Gte, nomad.app.v1.models.Gt, nomad.app.v1.models.Any_, nomad.app.v1.models.All, nomad.app.v1.models.None_]], nomad.app.v1.models.And, nomad.app.v1.models.Or, nomad.app.v1.models.Not] = None, user_id: str = None, **kwargs)¶ Uses the given painless script to update the entries by given query.
In most cases, the elasticsearch entry index should not be updated field by field; you should run index_all instead and fully replace documents from mongodb and archive files.
This method provides a faster direct method to update individual fiels, e.g. to quickly update fields for editing operations.
nomad.app¶
nomad.cli¶
Command line interface (CLI) for nomad. Provides a group/sub-command structure, think git, that offers various functionality to the command line user.
Use it from the command line with nomad --help or python -m nomad.cli --help to learn
more.
The CLI uses lazy_import for lazy loading modules. This has some limitations. You will
break lazy loading if an from x import y is used in the cli code. You will also
have to add imports via nomad.cli.lazy_import.lazy_module() before importing
them.
nomad.client¶
Install the NOMAD client library¶
The NOMAD client library is a Python module (part of the nomad Python package) that allows to access the NOMAD archive to retrieve and analyse (large amounts) of NOMAD’s archive data. It allows to use queries to filter for desired entries, bulk download the required parts of the respective archives, and navigate the results using NOMAD’s metainfo Python API.
To install the NOMAD Python package, you can use pip install to install our
source distribution
pip install nomad-lab
First example¶
'''
A simple example that uses the NOMAD client library to access the archive.
'''
from nomad.client import ArchiveQuery
from nomad.metainfo import units
query = ArchiveQuery(
# url='http://nomad-lab.eu/prod/rae/beta/api',
query={
'$and': {
'dft.code_name': 'VASP',
'$not': {
'atoms': ["Ti", "O"]
}
}
},
required={
'section_run': {
'section_single_configuration_calculation': {
'energy_total': '*'
},
'section_system': '*'
}
},
per_page=10,
max=None)
print(query)
# for i, result in enumerate(query):
# if i < 10:
# calc = result.section_run[0].section_single_configuration_calculation[-1]
# energy = calc.energy_total
# formula = calc.single_configuration_calculation_to_system_ref.chemical_composition_reduced
# print('%s: energy %s' % (formula, energy.to(units.hartree)))
This script should yield a result like this:
Number queries entries: 7628
Number of entries loaded in the last api call: 10
Bytes loaded in the last api call: 118048
Bytes loaded from this query: 118048
Number of downloaded entries: 10
Number of made api calls: 1
Cd2O2: energy -11467.827149010665 hartree
Sr2O2: energy -6551.45699684026 hartree
Sr2O2: energy -6551.461104765451 hartree
Be2O2: energy -178.6990610734937 hartree
Ca2O2: energy -1510.3938165430286 hartree
Ca2O2: energy -1510.3937761449583 hartree
Ba2O2: energy -16684.667362890417 hartree
Mg2O2: energy -548.9736595672932 hartree
Mg2O2: energy -548.9724185656775 hartree
Ca2O2: energy -1510.3908614326358 hartree
Let’s discuss the different elements here. First, we have a set of imports. The NOMAD source codes comes with various sub-modules. The client module contains everything related to what is described here; the metainfo is the Python interface to NOMAD’s common archive data format and its data type definitions; the config module simply contains configuration values (like the URL to the NOMAD API).
Next, we create an ArchiveQuery instance. This object will be responsible for talking
to NOMAD’s API for us in a transparent and lazy manner. This means, it will not download
all data right away, but do so when we are actually iterating through the results.
The archive query takes several parameters:
The
queryis a dictionary of search criteria. The query is used to filter all of NOMAD’s entry down to a set of desired entries. You can use NOMAD’s GUI to create queries and copy their Python equivalent with the<>-code button on the result list.The
requiredpart, allows to specify what parts of the archive should be downloaded. Leave it out to download the whole archives. Based on NOMAD’s Metainfo (the ‘schema’ of all archives), you can determine what sections to include and which to leave out. Here, we are interested in the first run (usually entries only have one run) and the first calculation result.With the optional
per_pageyou can determine, how many results are downloaded at a time. For bulk downloading many results, we recommend ~100. If you are just interested in the first results a lower number might increase performance.With the optional
max, we limit the maximum amount of entries that are downloaded, just to avoid accidentely iterating through a result set of unknown and potentially large size.
When you print the archive query object, you will get some basic statistics about the query and downloaded data.
The archive query object can be treated as a Python list-like. You use indices and ranges
to select results. Here we iterate through a slice and print the calculated energies
from the first calculation of the entries. Each result is a Python object with attributes
governed by the NOMAD Metainfo. Quantities yield numbers, string, or numpy arrays, while
sub-sections return lists of further objects. Here we navigate the sections section_run and
sub-section section_system to access the quantity energy_total. This quantity is a
number with an attached unit (Joule), which can be converted to something else (e.g. Hartree).
The create query object keeps all results in memory. Keep this in mind, when you are
accessing a large amount of query results. You should use ArchiveQuery.clear()
to remove unnecessary results.
The NOMAD Metainfo¶
You can imagine the NOMAD Metainfo as a complex schema for hiearchically organized scientific data. In this sense, the NOMAD Metainfo is a set of data type definitions. These definitions then govern how the archive for an data entry in NOMAD might look like. You can browse the hierarchy of definitions in our Metainfo browser.
Be aware, that the definitions entail everything that an entry could possibly contain, but not all entries contain all sections and all quantities. What an entry contains depends on the information that the respective uploaded data contained, what could be extracted, and of course what was calculated in the first place. To see what the archive of an concrete entry looks like, you can use the search interface, select an entry from the list fo search results, and click on the Archive tab.
To see inside an archive object in Python, you can use nomad.metainfo.MSection.m_to_dict()
which is provided by all archive objects. This will convert a (part of an) archive into a
regular, JSON-serializable Python dictionary.
For more details on the metainfo Python interface, consult the metainfo documentation.
The ArchiveQuery class¶
-
class
nomad.client.ArchiveQuery(query: dict = None, required: dict = None, url: str = None, username: str = None, password: str = None, parallel: int = 1, per_page: int = 10, max: int = 10000, raise_errors: bool = False, authentication: Union[Dict[str, str], nomad.client.KeycloakAuthenticator] = None)¶ Object of this class represent a query on the NOMAD Archive. It is solely configured through its constructor. After creation, it implements the Python
Sequenceinterface and therefore acts as a sequence of query results.Not all results are downloaded at once, expect that this class will continuesly pull results from the API, while you access or iterate to the far side of the result list.
-
query¶ A dictionary of search parameters. Consult the search API to get a comprehensive list of parameters.
-
required¶ A potentially nested dictionary of sections to retrieve.
-
url¶ Optional, override the default NOMAD API url.
-
username¶ Optional, allows authenticated access.
-
password¶ Optional, allows authenticated access.
-
per_page¶ Determine how many results are downloaded per page (or scroll window). Default is 10.
-
max¶ Optionally determine the maximum amount of downloaded archives. The iteration will stop if max is surpassed even if more results are available. Default is 10.000. None value will set it to unlimited.
-
raise_errors¶ There situations where archives for certain entries are unavailable. If set to True, this cases will raise an Exception. Otherwise, the entries with missing archives are simply skipped (default).
-
authentication¶ Optionally provide detailed authentication information. Usually, providing
usernameandpasswordshould suffice.
-
parallel¶ Number of processes to use to retrieve data in parallel. Only data from different uploads can be retrieved in parallel. Default is 1. The argument
per_pagewill refer to archived retrieved in one process per call.
-
__init__(query: dict = None, required: dict = None, url: str = None, username: str = None, password: str = None, parallel: int = 1, per_page: int = 10, max: int = 10000, raise_errors: bool = False, authentication: Union[Dict[str, str], nomad.client.KeycloakAuthenticator] = None)¶ Initialize self. See help(type(self)) for accurate signature.
-
Working with private data¶
Public NOMAD data can be accessed without any authentication; everyone can use our API
without the need for an account or login. However, if you want to work with your own
data that is not yet published, or embargoed data was shared with you, you need to
authenticate before accessing this data. Otherwise, you will simply not find it with
your queries. To authenticate simply provide your NOMAD username and password to the
ArchiveQuery constructor.
nomad.utils¶
Logging in nomad is structured. Structured logging means that log entries contain
dictionaries with quantities related to respective events. E.g. having the code,
parser, parser version, calc_id, mainfile, etc. for all events that happen during
calculation processing. This means the get_logger() and all logger functions
take keyword arguments for structured data. Otherwise get_logger() can
be used similar to the standard logging.getLogger.
Depending on the configuration all logs will also be send to a central logstash.
tests¶
The nomad@FAIRDI tests are based on the pytest library. Pytest uses fixtures to modularize setup and teardown of mocks, infrastructure, and other context objects. The following depicts the used hierarchy of fixtures:
Otherwise the test submodules follow the names of the nomad code modules.